1QZ4
 
 | |
1TRO
 
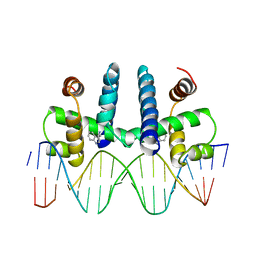 | | CRYSTAL STRUCTURE OF TRP REPRESSOR OPERATOR COMPLEX AT ATOMIC RESOLUTION | | 分子名称: | DNA (5'-D(*TP*GP*TP*AP*CP*TP*AP*GP*TP*TP*AP*AP*CP*TP*AP*GP*T P*AP*C)-3'), PROTEIN (TRP REPRESSOR), TRYPTOPHAN | | 著者 | Otwinowski, Z, Schevitz, R.W, Zhang, R.-G, Lawson, C.L, Joachimiak, A, Marmorstein, R, Luisi, B.F, Sigler, P.B. | | 登録日 | 1992-08-30 | | 公開日 | 1994-01-31 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of trp repressor/operator complex at atomic resolution.
Nature, 335, 1988
|
|
9EWK
 
 | | Solvent organization in ultrahigh-resolution protein crystal structure at room temperature | | 分子名称: | Crambin, ETHANOL | | 著者 | Chen, J.C.-H, Gilski, M, Chang, C, Borek, D, Rosenbaum, G, Lavens, A, Otwinowski, Z, Kubicki, M, Dauter, Z, Jaskolski, M, Joachimiak, A. | | 登録日 | 2024-04-04 | | 公開日 | 2024-09-04 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (0.7 Å) | | 主引用文献 | Solvent organization in the ultrahigh-resolution crystal structure of crambin at room temperature.
Iucrj, 11, 2024
|
|
1DW9
 
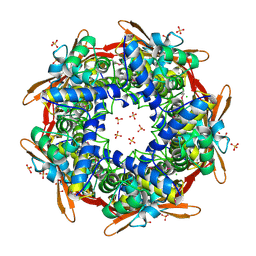 | | Structure of cyanase reveals that a novel dimeric and decameric arrangement of subunits is required for formation of the enzyme active site | | 分子名称: | CHLORIDE ION, CYANATE LYASE, SULFATE ION | | 著者 | Walsh, M.A, Otwinowski, Z, Perrakis, A, Anderson, P.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 1999-12-03 | | 公開日 | 2000-05-16 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structure of Cyanase Reveals that a Novel Dimeric and Decameric Arrangement of Subunits is Required for Formation of the Enzyme Active Site
Structure, 8, 2000
|
|
5DLL
 
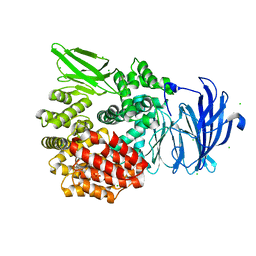 | | Aminopeptidase N (pepN) from Francisella tularensis subsp. tularensis SCHU S4 | | 分子名称: | Aminopeptidase N, CHLORIDE ION, PHOSPHATE ION, ... | | 著者 | Borek, D, Raczynska, J, Dubrovska, I, Grimshaw, S, Minasov, G, Shuvalova, L, Kwon, K, Anderson, W.F, Otwinowski, Z, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-09-07 | | 公開日 | 2015-09-16 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Aminopeptidase N (pepN) from Francisella tularensis subsp. tularensis SCHU S4
To Be Published
|
|
5DO7
 
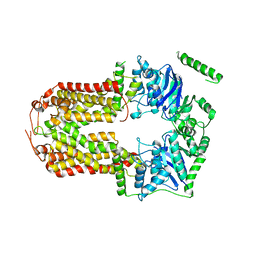 | | Crystal Structure of the Human Sterol Transporter ABCG5/ABCG8 | | 分子名称: | ATP-binding cassette sub-family G member 5, ATP-binding cassette sub-family G member 8 | | 著者 | Lee, J.-Y, Kinch, L.N, Borek, D.M, Urbatsch, I.L, Xie, X.-S, Grishin, N.V, Cohen, J.C, Otwinowski, Z, Hobbs, H.H, Rosenbaum, D.M. | | 登録日 | 2015-09-10 | | 公開日 | 2016-05-11 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (3.93 Å) | | 主引用文献 | Crystal structure of the human sterol transporter ABCG5/ABCG8.
Nature, 533, 2016
|
|
6ROA
 
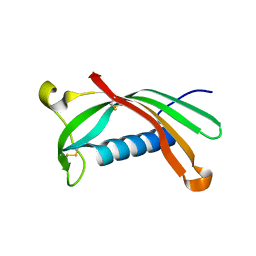 | | Crystal structure of V57G mutant of human cystatin C | | 分子名称: | Cystatin-C | | 著者 | Orlikowska, M, Behrendt, I, Borek, D, Otwinowski, Z, Skowron, P, Szymanska, A. | | 登録日 | 2019-05-10 | | 公開日 | 2019-08-07 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | NMR and crystallographic structural studies of the extremely stable monomeric variant of human cystatin C with single amino acid substitution.
Febs J., 287, 2020
|
|
4YPI
 
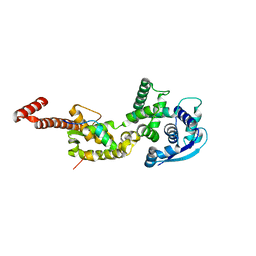 | | Structure of Ebola virus nucleoprotein N-terminal fragment bound to a peptide derived from Ebola VP35 | | 分子名称: | Nucleoprotein, Polymerase cofactor VP35 | | 著者 | Leung, D.W, Borek, D.M, Binning, J.M, Otwinowski, Z, Amarasinghe, G.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-03-13 | | 公開日 | 2015-04-08 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (3.71 Å) | | 主引用文献 | An Intrinsically Disordered Peptide from Ebola Virus VP35 Controls Viral RNA Synthesis by Modulating Nucleoprotein-RNA Interactions.
Cell Rep, 11, 2015
|
|
4X6Z
 
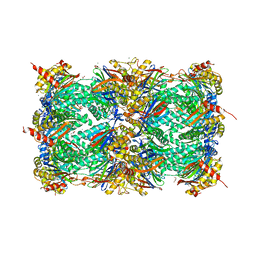 | | Yeast 20S proteasome in complex with PR-VI modulator | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Rostankowski, R, Witkowska, J, Borek, D, Otwinowski, Z, Jankowska, E. | | 登録日 | 2014-12-09 | | 公開日 | 2015-12-23 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structures revealed the common place of binding of low-molecular
mass activators with the 20S proteasome
To Be Published
|
|
5IZM
 
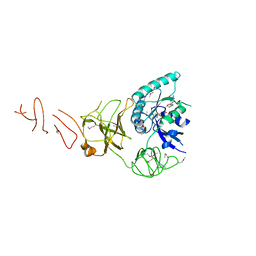 | |
1POC
 
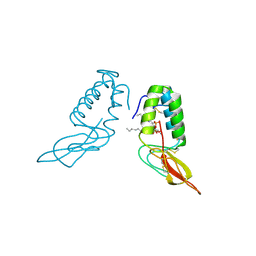 | |
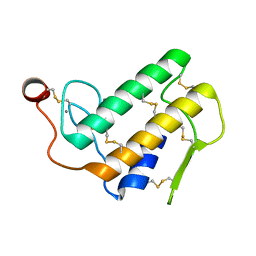
1POA
 
 | |
1POB
 
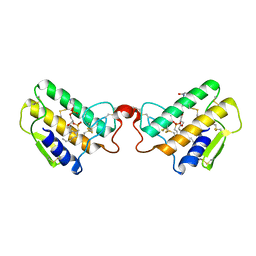 | | CRYSTAL STRUCTURE OF COBRA-VENOM PHOSPHOLIPASE A2 IN A COMPLEX WITH A TRANSITION-STATE ANALOGUE | | 分子名称: | 1-O-OCTYL-2-HEPTYLPHOSPHONYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, CALCIUM ION, PHOSPHOLIPASE A2 | | 著者 | White, S.P, Scott, D.L, Otwinowski, Z, Sigler, P.B. | | 登録日 | 1992-09-07 | | 公開日 | 1993-10-31 | | 最終更新日 | 2019-08-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of cobra-venom phospholipase A2 in a complex with a transition-state analogue.
Science, 250, 1990
|
|
5T8V
 
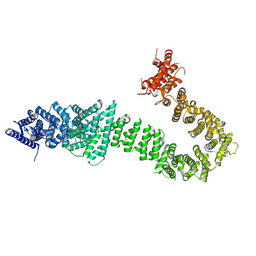 | | Chaetomium thermophilum cohesin loader SCC2, C-terminal fragment | | 分子名称: | CITRIC ACID, Putative uncharacterized protein | | 著者 | Tomchick, D.R, Yu, H, Kikuchi, S, Ouyang, Z, Borek, D, Otwinowski, Z. | | 登録日 | 2016-09-08 | | 公開日 | 2016-10-19 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.798 Å) | | 主引用文献 | Crystal structure of the cohesin loader Scc2 and insight into cohesinopathy.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6VSC
 
 | |
6VRS
 
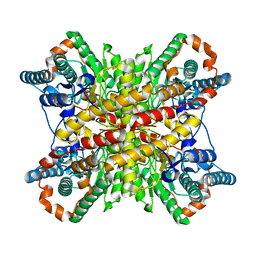 | | Single particle reconstruction of glucose isomerase from Streptomyces rubiginosus based on data acquired in the presence of substantial aberrations | | 分子名称: | MANGANESE (II) ION, xylose isomerase | | 著者 | Bromberg, R, Guo, Y, Borek, D, Otwinowski, Z. | | 登録日 | 2020-02-09 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | High-resolution cryo-EM reconstructions in the presence of substantial aberrations
Iucrj, 7, 2020
|
|
6VSA
 
 | |
4OFK
 
 | | Crystal Structure of SYG-2 D4 | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | 著者 | Ozkan, E, Borek, D, Otwinowski, Z, Garcia, K.C. | | 登録日 | 2014-01-15 | | 公開日 | 2014-02-19 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.802 Å) | | 主引用文献 | Extracellular Architecture of the SYG-1/SYG-2 Adhesion Complex Instructs Synaptogenesis.
Cell(Cambridge,Mass.), 156, 2014
|
|
4GHL
 
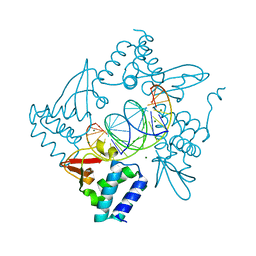 | | Structural Basis for Marburg virus VP35 mediate immune evasion mechanisms | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Ramanan, P, Borek, D.M, Otwinowski, Z, Leung, D.W, Amarasinghe, G.K. | | 登録日 | 2012-08-07 | | 公開日 | 2012-11-28 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Structural basis for Marburg virus VP35-mediated immune evasion mechanisms.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7U5T
 
 | |
7U5U
 
 | |
7U5S
 
 | |
5VR0
 
 | |
3L26
 
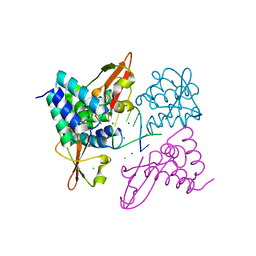 | | Crystal structure of Zaire Ebola VP35 interferon inhibitory domain bound to 8 bp dsRNA | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, Polymerase cofactor VP35, ... | | 著者 | Leung, D.W, Prins, K.C, Borek, D.M, Farahbakhsh, M, Tufariello, J.M, Ramanan, P, Nix, J.C, Helgeson, L.A, Otwinowski, Z, Honzatko, R.B, Basler, C.F, Amarasinghe, G.K. | | 登録日 | 2009-12-14 | | 公開日 | 2010-01-26 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3L27
 
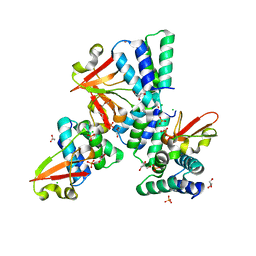 | | Crystal structure of Zaire Ebola VP35 interferon inhibitory domain R312A mutant | | 分子名称: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | 著者 | Leung, D.W, Prins, K.C, Borek, D.M, Farahbakhsh, M, Tufariello, J.M, Ramanan, P, Nix, J.C, Helgeson, L.A, Otwinowski, Z, Honzatko, R.B, Basler, C.F, Amarasinghe, G.K. | | 登録日 | 2009-12-14 | | 公開日 | 2010-01-26 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35.
Nat.Struct.Mol.Biol., 17, 2010
|
|
