4AF5
 
 | | Structure of the C. glutamicum AcnR Crystal Form I | | Descriptor: | CITRIC ACID, HTH-TYPE TRANSCRIPTIONAL REPRESSOR ACNR COMPND 2, MAGNESIUM ION | | Authors: | Garcia-Nafria, J, Baumgart, M, Turkenburg, J.P, Wilkinson, A.J, Bott, M, Wilson, K.S. | | Deposit date: | 2012-01-17 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal and Solution Studies Reveal that the Transcriptional Regulator Acnr of Corynebacterium Glutamicum is Regulated by Citrate:Mg2+ Binding to a Non-Canonical Pocket.
J.Biol.Chem., 288, 2013
|
|
4ACI
 
 | | Structure of the C. glutamicum AcnR Crystal Form II | | Descriptor: | CITRIC ACID, GLYCEROL, HTH-TYPE TRANSCRIPTIONAL REPRESSOR ACNR, ... | | Authors: | Garcia-Nafria, J, Baumgart, M, Turkenburg, J.P, Wilkinson, A.J, Bott, M, Wilson, K.S. | | Deposit date: | 2011-12-15 | | Release date: | 2012-12-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal and Solution Studies Reveal that the Transcriptional Regulator Acnr of Corynebacterium Glutamicum is Regulated by Citrate:Mg2+ Binding to a Non-Canonical Pocket.
J.Biol.Chem., 288, 2013
|
|
7QFX
 
 | | Crystal structure of Old Yellow Enzyme AnOYE8 from Aspergillus niger | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FLAVIN MONONUCLEOTIDE, NADH-dependent flavin oxidoreductase, ... | | Authors: | Robescu, M.S, Loprete, G, Vascon, F, Gasparotto, M, Filippini, F, Bergantino, E, Cendron, L. | | Deposit date: | 2021-12-06 | | Release date: | 2022-04-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Family Keeps on Growing: Four Novel Fungal OYEs Characterized.
Int J Mol Sci, 23, 2022
|
|
1ZTO
 
 | | INACTIVATION GATE OF POTASSIUM CHANNEL RCK4, NMR, 8 STRUCTURES | | Descriptor: | POTASSIUM CHANNEL PROTEIN RCK4 | | Authors: | Antz, C, Geyer, M, Fakler, B, Schott, M, Frank, R, Guy, H.R, Ruppersberg, J.P, Kalbitzer, H.R. | | Deposit date: | 1996-11-15 | | Release date: | 1997-06-05 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR structure of inactivation gates from mammalian voltage-dependent potassium channels.
Nature, 385, 1997
|
|
3T41
 
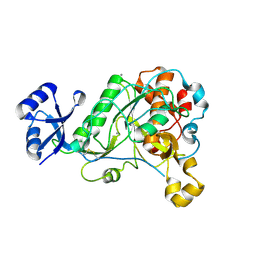 | | 1.95 Angstrom Resolution Crystal Structure of Epidermin Leader Peptide Processing Serine Protease (EpiP) S393A Mutant from Staphylococcus aureus | | Descriptor: | CALCIUM ION, CHLORIDE ION, Epidermin leader peptide processing serine protease EpiP | | Authors: | Minasov, G, Kuhn, M, Ruan, J, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-25 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Resolution Crystal Structure of Epidermin Leader Peptide Processing Serine Protease (EpiP) S393A Mutant from Staphylococcus aureus.
TO BE PUBLISHED
|
|
1ZTN
 
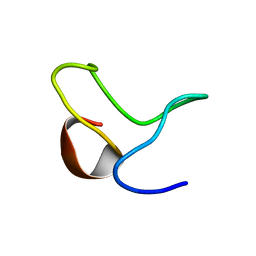 | | INACTIVATION GATE OF POTASSIUM CHANNEL RAW3, NMR, 8 STRUCTURES | | Descriptor: | Potassium voltage-gated channel subfamily C member 4 | | Authors: | Antz, C, Geyer, M, Fakler, B, Schott, M, Frank, R, Guy, H.R, Ruppersberg, J.P, Kalbitzer, H.R. | | Deposit date: | 1996-11-15 | | Release date: | 1997-06-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of inactivation gates from mammalian voltage-dependent potassium channels.
Nature, 385, 1997
|
|
6AXD
 
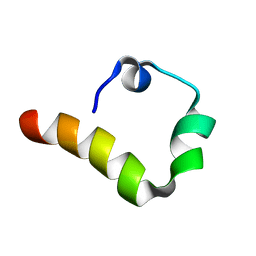 | |
3VK1
 
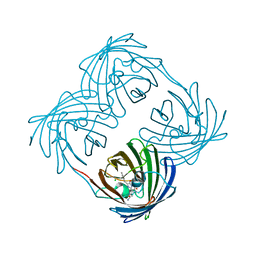 | | Green-fluorescent variant of the non-fluorescent chromoprotein Rtms5 | | Descriptor: | CHLORIDE ION, GFP-like non-fluorescent chromoprotein, IODIDE ION | | Authors: | Battad, J.M, Traore, D.A.K, Wilce, M, Byres, M, Rossjohn, J, Devenish, R.J, Prescott, M. | | Deposit date: | 2011-11-07 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Green Fluorescent Protein Containing a QFG Tri-Peptide Chromophore: Optical Properties and X-Ray Crystal Structure.
Plos One, 7, 2012
|
|
2K0L
 
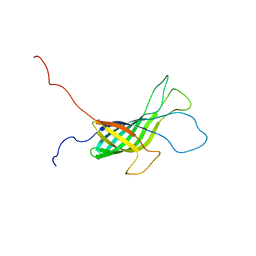 | | NMR structure of the transmembrane domain of the Outer Membrane Protein A from Klebsiella pneumoniae in DHPC micelles. | | Descriptor: | Outer membrane protein A | | Authors: | Renault, M, Saurel, O, Gervais, V, Lohr, F, Reat, V, Piotto, M, Milon, A. | | Deposit date: | 2008-02-04 | | Release date: | 2008-12-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution state NMR structure and dynamics of KpOmpA, a 210 residue transmembrane domain possessing a high potential for immunological applications.
J.Mol.Biol., 385, 2009
|
|
4AC6
 
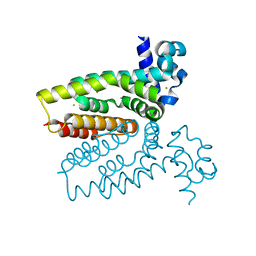 | | Corynebacterium glutamicum AcnR AU derivative structure | | Descriptor: | GOLD ION, HTH-TYPE TRANSCRIPTIONAL REPRESSOR ACNR | | Authors: | Garcia-Nafria, J, Baumgart, M, Turkenburg, J.P, Wilkinson, A.J, Bott, M, Wilson, K.S. | | Deposit date: | 2011-12-14 | | Release date: | 2012-12-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal and Solution Studies Reveal that the Transcriptional Regulator Acnr of Corynebacterium Glutamicum is Regulated by Citrate:Mg2+ Binding to a Non-Canonical Pocket.
J.Biol.Chem., 288, 2013
|
|
2JW6
 
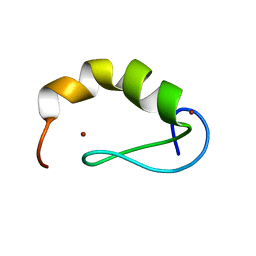 | | Solution structure of the DEAF1 MYND domain | | Descriptor: | Deformed epidermal autoregulatory factor 1 homolog, ZINC ION | | Authors: | Spadaccini, R, Perrin, H, Bottomley, M, Ansieu, S, Sattler, M. | | Deposit date: | 2007-10-08 | | Release date: | 2007-12-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Retraction notice to "Structure and functional analysis of the MYND domain" [J. Mol. Biol. (2006) 358, 498-508].
J.Mol.Biol., 376, 2008
|
|
3F51
 
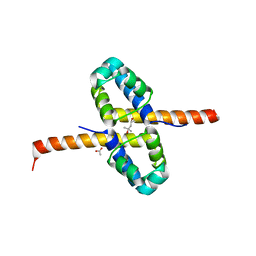 | | Crystal Structure of the clp gene regulator ClgR from Corynebacterium glutamicum | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Clp gene regulator (ClgR) | | Authors: | Russo, S, Schweitzer, J.E, Polen, T, Bott, M, Pohl, E. | | Deposit date: | 2008-11-03 | | Release date: | 2008-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the caseinolytic protease gene regulator, a transcriptional activator in actinomycetes
J.Biol.Chem., 284, 2009
|
|
4A24
 
 | | Structural and functional analysis of the DEAF-1 and BS69 MYND domains | | Descriptor: | DEFORMED EPIDERMAL AUTOREGULATORY FACTOR 1 HOMOLOG, ZINC ION | | Authors: | Kateb, F, Perrin, H, Tripsianes, K, Zou, P, Spadaccini, R, Bottomley, M, Bepperling, A, Ansieau, S, Sattler, M. | | Deposit date: | 2011-09-22 | | Release date: | 2012-11-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Analysis of the Deaf-1 and Bs69 Mynd Domains.
Plos One, 8, 2013
|
|
1OVD
 
 | | THE K136E MUTANT OF LACTOCOCCUS LACTIS DIHYDROOROTATE DEHYDROGENASE A IN COMPLEX WITH OROTATE | | Descriptor: | DIHYDROOROTATE DEHYDROGENASE A, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Norager, S, Arent, S, Bjornberg, O, Ottosen, M, Lo Leggio, L, Jensen, K.F, Larsen, S. | | Deposit date: | 2003-03-26 | | Release date: | 2003-08-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function.
J.Biol.Chem., 278, 2003
|
|
3F52
 
 | | Crystal structure of the clp gene regulator ClgR from C. glutamicum | | Descriptor: | GLYCEROL, clp gene regulator (ClgR) | | Authors: | Russo, S, Schweitzer, J.E, Polen, T, Bott, M, Pohl, E. | | Deposit date: | 2008-11-03 | | Release date: | 2008-11-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the caseinolytic protease gene regulator, a transcriptional activator in actinomycetes
J.Biol.Chem., 284, 2009
|
|
6GPH
 
 | | Structure of human Heat shock protein 90-alpha N-terminal domain (Hsp90-NTD) variant K112A in complex with AMPCP | | Descriptor: | Heat shock protein HSP 90-alpha, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER | | Authors: | Tassone, G, Pozzi, C, Mangani, S, Botta, M. | | Deposit date: | 2018-06-05 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Probing the role of Arg97 in Heat shock protein 90 N-terminal domain from the parasite Leishmania braziliensis through site-directed mutagenesis on the human counterpart.
Biochim Biophys Acta Proteins Proteom, 1866, 2018
|
|
6GPT
 
 | | Structure of human Heat shock protein 90-alpha N-terminal domain (Hsp90-NTD) in complex with AMPCP | | Descriptor: | Heat shock protein HSP 90-alpha, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER | | Authors: | Tassone, G, Pozzi, C, Mangani, S, Botta, M. | | Deposit date: | 2018-06-07 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the role of Arg97 in Heat shock protein 90 N-terminal domain from the parasite Leishmania braziliensis through site-directed mutagenesis on the human counterpart.
Biochim Biophys Acta Proteins Proteom, 1866, 2018
|
|
6GQR
 
 | | Structure of human Heat shock protein 90-alpha N-terminal domain (Hsp90-NTD) in complex with AMPCPP | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, Heat shock protein HSP 90-alpha, MAGNESIUM ION | | Authors: | Tassone, G, Pozzi, C, Mangani, S, Botta, M. | | Deposit date: | 2018-06-08 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Probing the role of Arg97 in Heat shock protein 90 N-terminal domain from the parasite Leishmania braziliensis through site-directed mutagenesis on the human counterpart.
Biochim Biophys Acta Proteins Proteom, 1866, 2018
|
|
6GQU
 
 | | Structure of human Heat shock protein 90-alpha N-terminal domain (Hsp90-NTD) variant K112R in complex with cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Heat shock protein HSP 90-alpha | | Authors: | Tassone, G, Pozzi, C, Mangani, S, Botta, M. | | Deposit date: | 2018-06-08 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Probing the role of Arg97 in Heat shock protein 90 N-terminal domain from the parasite Leishmania braziliensis through site-directed mutagenesis on the human counterpart.
Biochim Biophys Acta Proteins Proteom, 1866, 2018
|
|
6GR4
 
 | | Structure of human Heat shock protein 90-alpha N-terminal domain (Hsp90-NTD) variant K112R | | Descriptor: | CHLORIDE ION, Heat shock protein HSP 90-alpha, MAGNESIUM ION | | Authors: | Tassone, G, Pozzi, C, Mangani, S, Botta, M. | | Deposit date: | 2018-06-08 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Probing the role of Arg97 in Heat shock protein 90 N-terminal domain from the parasite Leishmania braziliensis through site-directed mutagenesis on the human counterpart.
Biochim Biophys Acta Proteins Proteom, 1866, 2018
|
|
6GPR
 
 | | Structure of human Heat shock protein 90-alpha N-terminal domain (Hsp90-NTD) in complex with cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Heat shock protein HSP 90-alpha | | Authors: | Tassone, G, Pozzi, C, Mangani, S, Botta, M. | | Deposit date: | 2018-06-06 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Probing the role of Arg97 in Heat shock protein 90 N-terminal domain from the parasite Leishmania braziliensis through site-directed mutagenesis on the human counterpart.
Biochim Biophys Acta Proteins Proteom, 1866, 2018
|
|
6GQS
 
 | | Structure of human Heat shock protein 90-alpha N-terminal domain (Hsp90-NTD) variant K112R in complex with AMPCP | | Descriptor: | Heat shock protein HSP 90-alpha, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER | | Authors: | Tassone, G, Pozzi, C, Mangani, S, Botta, M. | | Deposit date: | 2018-06-08 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Probing the role of Arg97 in Heat shock protein 90 N-terminal domain from the parasite Leishmania braziliensis through site-directed mutagenesis on the human counterpart.
Biochim Biophys Acta Proteins Proteom, 1866, 2018
|
|
6GPW
 
 | | Structure of human Heat shock protein 90-alpha N-terminal domain (Hsp90-NTD) | | Descriptor: | CHLORIDE ION, Heat shock protein HSP 90-alpha, MAGNESIUM ION | | Authors: | Tassone, G, Pozzi, C, Mangani, S, Botta, M. | | Deposit date: | 2018-06-07 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Probing the role of Arg97 in Heat shock protein 90 N-terminal domain from the parasite Leishmania braziliensis through site-directed mutagenesis on the human counterpart.
Biochim Biophys Acta Proteins Proteom, 1866, 2018
|
|
6GR1
 
 | | Structure of human Heat shock protein 90-alpha N-terminal domain (Hsp90-NTD) variant K112R in complex with AMPCPP | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, Heat shock protein HSP 90-alpha, MAGNESIUM ION | | Authors: | Tassone, G, Pozzi, C, Mangani, S, Botta, M. | | Deposit date: | 2018-06-08 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Probing the role of Arg97 in Heat shock protein 90 N-terminal domain from the parasite Leishmania braziliensis through site-directed mutagenesis on the human counterpart.
Biochim Biophys Acta Proteins Proteom, 1866, 2018
|
|
6GR5
 
 | | Structure of human Heat shock protein 90-alpha N-terminal domain (Hsp90-NTD) variant K112R in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Heat shock protein HSP 90-alpha, MAGNESIUM ION | | Authors: | Tassone, G, Pozzi, C, Mangani, S, Botta, M. | | Deposit date: | 2018-06-08 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Probing the role of Arg97 in Heat shock protein 90 N-terminal domain from the parasite Leishmania braziliensis through site-directed mutagenesis on the human counterpart.
Biochim Biophys Acta Proteins Proteom, 1866, 2018
|
|
