3ZWM
 
 | | Crystal structure of ADP ribosyl cyclase complexed with substrate NAD and product cADPR | | Descriptor: | ADP-RIBOSYL CYLCASE, CYCLIC ADENOSINE DIPHOSPHATE-RIBOSE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kotaka, M, Graeff, R, Zhang, L.H, Lee, H.C, Hao, Q. | | Deposit date: | 2011-08-02 | | Release date: | 2011-11-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Studies of Intermediates Along the Cyclization Pathway of Aplysia Adp-Ribosyl Cyclase.
J.Mol.Biol., 415, 2012
|
|
3ZWO
 
 | | Crystal structure of ADP ribosyl cyclase complexed with reaction intermediate | | Descriptor: | 3-(AMINOCARBONYL)-1-[(2R,3R,4S,5R)-5-({[(S)-{[(S)-{[(2R,3S,4R,5R)-5-(2-AMINO-6-OXO-1,6-DIHYDRO-9H-PURIN-9-YL)-3,4-DIHYD ROXYTETRAHYDROFURAN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]OXY}METHYL)-3,4-DIHYDROXYTETRAHYDROFURAN-2- YL]PYRIDINIUM, ADP-RIBOSYL CYCLASE, GUANOSINE DIPHOSPHATE RIBOSE | | Authors: | Kotaka, M, Graeff, R, Zhang, L.H, Lee, H.C, Hao, Q. | | Deposit date: | 2011-08-02 | | Release date: | 2011-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Studies of Intermediates Along the Cyclization Pathway of Aplysia Adp-Ribosyl Cyclase.
J.Mol.Biol., 415, 2012
|
|
3ZWY
 
 | | Crystal structure of ADP-ribosyl cyclase complexed with 8-bromo-ADP- ribose and cyclic 8-bromo-cyclic-ADP-ribose | | Descriptor: | (2R,3R,4S,5R,13R,14S,15R,16R)-24-amino-18-bromo-3,4,14,15-tetrahydroxy-7,9,11,25,26-pentaoxa-17,19,22-triaza-1-azonia-8 ,10-diphosphapentacyclo[18.3.1.1^2,5^.1^13,16^.0^17,21^]hexacosa-1(24),18,20,22-tetraene-8,10-diolate 8,10-dioxide, ADP-RIBOSYL CYCLASE, [(2R,3S,4R,5R)-5-(6-amino-8-bromo-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3S,4S)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate | | Authors: | Kotaka, M, Graeff, R, Zhang, L.H, Lee, H.C, Hao, Q. | | Deposit date: | 2011-08-03 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Studies of Intermediates Along the Cyclization Pathway of Aplysia Adp-Ribosyl Cyclase.
To be Published
|
|
3ZWX
 
 | | Crystal structure of ADP-ribosyl cyclase complexed with 8-bromo-ADP- ribose | | Descriptor: | ADP-RIBOSYL CYCLASE, CHLORIDE ION, [(2R,3S,4R,5R)-5-(6-amino-8-bromo-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3S,4S)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate | | Authors: | Kotaka, M, Graeff, R, Zhang, L.H, Lee, H.C, Hao, Q. | | Deposit date: | 2011-08-03 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Studies of Intermediates Along the Cyclization Pathway of Aplysia Adp-Ribosyl Cyclase.
To be Published
|
|
3ZWW
 
 | | Crystal structure of ADP-ribosyl cyclase complexed with ara-2'F-ADP- ribose at 2.3 angstrom | | Descriptor: | ADP-RIBOSYL CYCLASE, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3R,4R)-4-fluoro-3-hydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate | | Authors: | Kotaka, M, Graeff, R, Zhang, L.H, Lee, H.C, Hao, Q. | | Deposit date: | 2011-08-03 | | Release date: | 2011-11-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Studies of Intermediates Along the Cyclization Pathway of Aplysia Adp-Ribosyl Cyclase.
J.Mol.Biol., 415, 2012
|
|
3ZWN
 
 | | Crystal structure of Aplysia cyclase complexed with substrate NGD and product cGDPR | | Descriptor: | 3-(AMINOCARBONYL)-1-[(2R,3R,4S,5R)-5-({[(S)-{[(S)-{[(2R,3S,4R,5R)-5-(2-AMINO-6-OXO-1,6-DIHYDRO-9H-PURIN-9-YL)-3,4-DIHYD ROXYTETRAHYDROFURAN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]OXY}METHYL)-3,4-DIHYDROXYTETRAHYDROFURAN-2- YL]PYRIDINIUM, ADP-RIBOSYL CYCLASE, CYCLIC GUANOSINE DIPHOSPHATE-RIBOSE | | Authors: | Kotaka, M, Graeff, R, Zhang, L.H, Lee, H.C, Hao, Q. | | Deposit date: | 2011-08-02 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Studies of Intermediates Along the Cyclization Pathway of Aplysia Adp-Ribosyl Cyclase.
J.Mol.Biol., 415, 2012
|
|
3ZWV
 
 | | Crystal structure of ADP-ribosyl cyclase complexed with ara-2'F-ADP- ribose at 2.3 angstrom | | Descriptor: | ADP-RIBOSYL CYCLASE, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3R,4R)-4-fluoro-3-hydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate | | Authors: | Kotaka, M, Graeff, R, Zhang, L.H, Lee, H.C, Hao, Q. | | Deposit date: | 2011-08-03 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural studies of intermediates along the cyclization pathway of Aplysia ADP-ribosyl cyclase.
J. Mol. Biol., 415, 2012
|
|
3ZWP
 
 | | Crystal structure of ADP ribosyl cyclase complexed with ara-2'F-ADP- ribose at 2.1 angstrom | | Descriptor: | ADP-RIBOSYL CYCLASE, GLYCEROL, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3R,4R)-4-fluoro-3-hydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate | | Authors: | Kotaka, M, Graeff, R, Zhang, L.H, Lee, H.C, Hao, Q. | | Deposit date: | 2011-08-02 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural Studies of Intermediates Along the Cyclization Pathway of Aplysia Adp-Ribosyl Cyclase.
J.Mol.Biol., 415, 2012
|
|
8BQ1
 
 | | Herpes simplex virus type 1 protease | | Descriptor: | Assemblin, GLYCEROL | | Authors: | Pachota, M, Grzywa, R, Plewka, J, Wilk, P, Mackereth, C, Czarna, A, Sienczyk, M, Pyrc, K. | | Deposit date: | 2022-11-18 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Herpes simplex virus type 1 protease
To Be Published
|
|
6JJN
 
 | | Crystal structure of Mumps virus hemagglutinin-neuraminidase bound to sialyl lewisX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HN protein, ... | | Authors: | Kubota, M, Matsuoka, R, Suzuki, T, Yonekura, K, Yanagi, Y, Hashiguchi, T. | | Deposit date: | 2019-02-26 | | Release date: | 2019-05-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Mechanism of the Flexible Glycan Receptor Recognition by Mumps Virus.
J.Virol., 93, 2019
|
|
6JJM
 
 | | Crystal structure of Mumps virus hemagglutinin-neuraminidase bound to the oligosaccharide portion of the GM2 ganglioside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HN protein, ... | | Authors: | Kubota, M, Matsuoka, R, Suzuki, T, Yonekura, K, Yanagi, Y, Hashiguchi, T. | | Deposit date: | 2019-02-26 | | Release date: | 2019-05-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Molecular Mechanism of the Flexible Glycan Receptor Recognition by Mumps Virus.
J.Virol., 93, 2019
|
|
5B2C
 
 | | Crystal structure of Mumps virus hemagglutinin-neuraminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HN protein, SULFATE ION | | Authors: | Kubota, M, Takeuchi, K, Watanabe, S, Ohno, S, Matsuoka, R, Kohda, D, Hiramatsu, H, Suzuki, Y, Nakayama, T, Terada, T, Shimizu, K, Shimizu, N, Yanagi, Y, Hashiguchi, T. | | Deposit date: | 2016-01-14 | | Release date: | 2016-09-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.238 Å) | | Cite: | Trisaccharide containing alpha 2,3-linked sialic acid is a receptor for mumps virus
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5B2D
 
 | | Crystal structure of Mumps virus hemagglutinin-neuraminidase bound to 3-sialyllactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HN protein, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Kubota, M, Takeuchi, K, Watanabe, S, Ohno, S, Matsuoka, R, Kohda, D, Hiramatsu, H, Suzuki, Y, Nakayama, T, Terada, T, Shimizu, K, Shimizu, N, Yanagi, Y, Hashiguchi, T. | | Deposit date: | 2016-01-14 | | Release date: | 2016-09-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.177 Å) | | Cite: | Trisaccharide containing alpha 2,3-linked sialic acid is a receptor for mumps virus
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2J0W
 
 | | Crystal structure of E. coli aspartokinase III in complex with aspartate and ADP (R-state) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ASPARTIC ACID, CHLORIDE ION, ... | | Authors: | Kotaka, M, Ren, J, Lockyer, M, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2006-08-07 | | Release date: | 2006-08-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of R- and T-State Escherichia Coli Aspartokinase III: Mechanisms of the Allosteric Transition and Inhibition by Lysine.
J.Biol.Chem., 281, 2006
|
|
2J0X
 
 | | CRYSTAL STRUCTURE OF E. COLI ASPARTOKINASE III IN COMPLEX WITH LYSINE AND ASPARTATE (T-STATE) | | Descriptor: | ASPARTIC ACID, LYSINE, LYSINE-SENSITIVE ASPARTOKINASE 3, ... | | Authors: | Kotaka, M, Ren, J, Lockyer, M, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2006-08-07 | | Release date: | 2006-08-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of R- and T-State Escherichia Coli Aspartokinase III: Mechanisms of the Allosteric Transition and Inhibition by Lysine.
J.Biol.Chem., 281, 2006
|
|
2VUS
 
 | | Crystal structure of unliganded NmrA-AreA zinc finger complex | | Descriptor: | CHLORIDE ION, NITROGEN METABOLITE REPRESSION REGULATOR NMRA, NITROGEN REGULATORY PROTEIN AREA, ... | | Authors: | Kotaka, M, Johnson, C, Lamb, H.K, Hawkins, A.R, Ren, J, Stammers, D.K. | | Deposit date: | 2008-05-30 | | Release date: | 2008-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Analysis of the Recognition of the Negative Regulator Nmra and DNA by the Zinc Finger from the Gata-Type Transcription Factor Area.
J.Mol.Biol., 381, 2008
|
|
2VUU
 
 | | Crystal structure of NADP-bound NmrA-AreA zinc finger complex | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NITROGEN METABOLITE REPRESSION REGULATOR NMRA, NITROGEN REGULATORY PROTEIN AREA, ... | | Authors: | Kotaka, M, Johnson, C, Lamb, H.K, Hawkins, A.R, Ren, J, Stammers, D.K. | | Deposit date: | 2008-05-30 | | Release date: | 2008-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Analysis of the Recognition of the Negative Regulator Nmra and DNA by the Zinc Finger from the Gata-Type Transcription Factor Area.
J.Mol.Biol., 381, 2008
|
|
2VUT
 
 | | Crystal structure of NAD-bound NmrA-AreA zinc finger complex | | Descriptor: | CHLORIDE ION, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Kotaka, M, Johnson, C, Lamb, H.K, Hawkins, A.R, Ren, J, Stammers, D.K. | | Deposit date: | 2008-05-30 | | Release date: | 2008-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Analysis of the Recognition of the Negative Regulator Nmra and DNA by the Zinc Finger from the Gata-Type Transcription Factor Area.
J.Mol.Biol., 381, 2008
|
|
2W3X
 
 | | Crystal structure of a bifunctional hotdog fold thioesterase in enediyne biosynthesis, CalE7 | | Descriptor: | CALE7, GLYCEROL, O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500), ... | | Authors: | Kotaka, M, Kong, R, Qureshi, I, Ho, Q.S, Sun, H, Liew, C.W, Goh, L.P, Cheung, P, Mu, Y, Lescar, J, Liang, Z.X. | | Deposit date: | 2008-11-17 | | Release date: | 2009-04-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and Catalytic Mechanism of the Thioesterase Cale7 in Enediyne Biosynthesis.
J.Biol.Chem., 284, 2009
|
|
2VN1
 
 | | Crystal structure of the FK506-binding domain of Plasmodium falciparum FKBP35 in complex with FK506 | | Descriptor: | 70 KDA PEPTIDYLPROLYL ISOMERASE, 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN | | Authors: | Kotaka, M, Alag, R, Ye, H, Preiser, P.R, Yoon, H.S, Lescar, J. | | Deposit date: | 2008-01-30 | | Release date: | 2008-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of the Fk506 Binding Domain of Plasmodium Falciparum Fkbp35 in Complex with Fk506.
Biochemistry, 47, 2008
|
|
3A28
 
 | | Crystal structure of L-2,3-butanediol dehydrogenase | | Descriptor: | BETA-MERCAPTOETHANOL, L-2.3-butanediol dehydrogenase, MAGNESIUM ION, ... | | Authors: | Otagiri, M, Kurisu, G, Ui, S, Kusunoki, M. | | Deposit date: | 2009-05-02 | | Release date: | 2009-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for chiral substrate recognition by two 2,3-butanediol dehydrogenases
Febs Lett., 584, 2010
|
|
2CCG
 
 | | Crystal structure of His-tagged S. aureus thymidylate kinase complexed with thymidine monophosphate (TMP) | | Descriptor: | THYMIDINE-5'-PHOSPHATE, THYMIDYLATE KINASE | | Authors: | Kotaka, M, Dhaliwal, B, Ren, J, Nichols, C.E, Angell, R, Lockyer, M, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2006-01-16 | | Release date: | 2006-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of S. Aureus Thymidylate Kinase Reveal an Atypical Active Site Configuration and an Intermediate Conformational State Upon Substrate Binding
Protein Sci., 15, 2006
|
|
2CCK
 
 | | CRYSTAL STRUCTURE OF UNLIGANDED S. AUREUS THYMIDYLATE KINASE | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, THYMIDYLATE KINASE | | Authors: | Kotaka, M, Dhaliwal, B, Ren, J, Nichols, C.E, Angell, R, Lockyer, M, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2006-01-16 | | Release date: | 2006-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structures of S. Aureus Thymidylate Kinase Reveal an Atypical Active Site Configuration and an Intermediate Conformational State Upon Substrate Binding
Protein Sci., 15, 2006
|
|
2CCJ
 
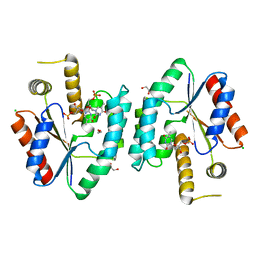 | | Crystal structure of S. aureus thymidylate kinase complexed with thymidine monophosphate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, THYMIDINE-5'-PHOSPHATE, ... | | Authors: | Kotaka, M, Dhaliwal, B, Ren, J, Nichols, C.E, Angell, R, Lockyer, M, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2006-01-16 | | Release date: | 2006-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of S. Aureus Thymidylate Kinase Reveal an Atypical Active Site Configuration and an Intermediate Conformational State Upon Substrate Binding
Protein Sci., 15, 2006
|
|
2BHL
 
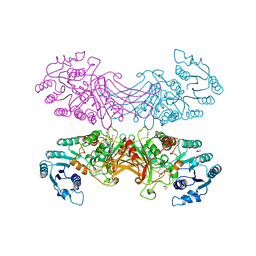 | | X-RAY STRUCTURE OF HUMAN GLUCOSE-6-PHOSPHATE DEHYDROGENASE (DELETION VARIANT) COMPLEXED WITH GLUCOSE-6-PHOSPHATE | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, GLUCOSE-6-PHOSPHATE 1-DEHYDROGENASE, GLYCEROL | | Authors: | Kotaka, M, Gover, S, Lam, V.M.S, Adams, M.J. | | Deposit date: | 2005-01-13 | | Release date: | 2005-04-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Studies of Glucose-6-Phosphate and Nadp+ Binding to Human Glucose-6-Phosphate Dehydrogenase
Acta Crystallogr.,Sect.D, 61, 2005
|
|
