3T9H
 
 | | Kainate bound to a double cysteine mutant (A452C/S652C) of the ligand binding domain of GluA2 | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 2, ZINC ION | | Authors: | Ahmed, A.H, Wang, S, Chuang, H.H, Oswald, R.E. | | Deposit date: | 2011-08-02 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Mechanism of AMPA Receptor Activation by Partial Agonists: DISULFIDE TRAPPING OF CLOSED LOBE CONFORMATIONS.
J.Biol.Chem., 286, 2011
|
|
3T9V
 
 | |
3T9X
 
 | |
3T93
 
 | |
3DP6
 
 | | Crystal structure of the binding domain of the AMPA subunit GluR2 bound to glutamate | | Descriptor: | GLUTAMIC ACID, Glutamate receptor 2, ZINC ION | | Authors: | Ahmed, A.H, Wang, Q, Sondermann, H, Oswald, R.E. | | Deposit date: | 2008-07-07 | | Release date: | 2008-11-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of the S1S2 glutamate binding domain of GLuR3.
Proteins, 75, 2008
|
|
3DLN
 
 | | Crystal structure of the binding domain of the AMPA subunit GluR3 bound to glutamate | | Descriptor: | GLUTAMIC ACID, Glutamate receptor 3, ZINC ION | | Authors: | Ahmed, A.H, Wang, Q, Sondermann, H, Oswald, R.E. | | Deposit date: | 2008-06-27 | | Release date: | 2008-11-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure of the S1S2 glutamate binding domain of GLuR3.
Proteins, 75, 2008
|
|
3DP4
 
 | | Crystal structure of the binding domain of the AMPA subunit GluR3 bound to AMPA | | Descriptor: | (S)-ALPHA-AMINO-3-HYDROXY-5-METHYL-4-ISOXAZOLEPROPIONIC ACID, Glutamate receptor 3, ZINC ION | | Authors: | Ahmed, A.H, Wang, Q, Sondermann, H, Oswald, R.E. | | Deposit date: | 2008-07-07 | | Release date: | 2008-11-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure of the S1S2 glutamate binding domain of GLuR3.
Proteins, 75, 2008
|
|
3H06
 
 | |
3H03
 
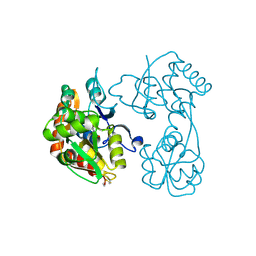 | |
3IL1
 
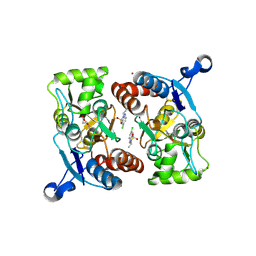 | | Crystal structure of the AMPA subunit GluR2 bound to the allosteric modulator, IDRA-21 | | Descriptor: | (3S)-7-chloro-3-methyl-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide, GLUTAMIC ACID, Glutamate receptor 2, ... | | Authors: | Ahmed, A.H, Ptak, C.P, Oswald, R.E. | | Deposit date: | 2009-08-06 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Probing the allosteric modulator binding site of GluR2 with thiazide derivatives
Biochemistry, 48, 2009
|
|
3ILT
 
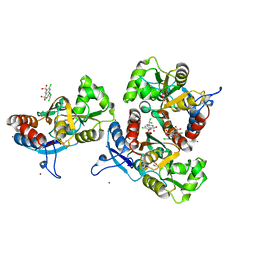 | | Crystal structure of the AMPA subunit GluR2 bound to the allosteric modulator, trichlormethiazide | | Descriptor: | 6-CHLORO-3-(DICHLOROMETHYL)-3,4-DIHYDRO-2H-1,2,4-BENZOTHIADIAZINE-7-SULFONAMIDE 1,1-DIOXIDE, GLUTAMIC ACID, Glutamate receptor 2, ... | | Authors: | Ahmed, A.H, Ptak, C.P, Oswald, R.E. | | Deposit date: | 2009-08-07 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.107 Å) | | Cite: | Probing the allosteric modulator binding site of GluR2 with thiazide derivatives
Biochemistry, 48, 2009
|
|
3IK6
 
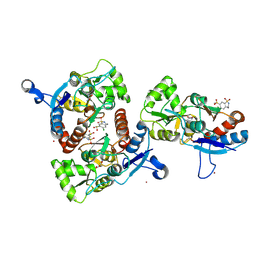 | | Crystal structure of the AMPA subunit GluR2 bound to the allosteric modulator, chlorothiazide | | Descriptor: | 6-chloro-3,4-dihydro-2H-1,2,4-benzothiadiazine-7-sulfonamide 1,1-dioxide, GLUTAMIC ACID, Glutamate receptor 2, ... | | Authors: | Ptak, C.P, Ahmed, A.H, Oswald, R.E. | | Deposit date: | 2009-08-05 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Probing the allosteric modulator binding site of GluR2 with thiazide derivatives
Biochemistry, 48, 2009
|
|
3IJO
 
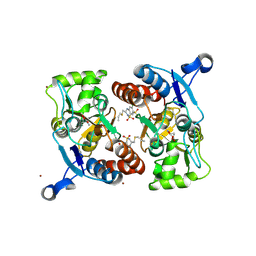 | | Crystal structure of the AMPA subunit GluR2 bound to the allosteric modulator, althiazide | | Descriptor: | (3S)-6-chloro-3-[(prop-2-en-1-ylsulfanyl)methyl]-3,4-dihydro-2H-1,2,4-benzothiadiazine-7-sulfonamide 1,1-dioxide, GLUTAMIC ACID, Glutamate receptor 2, ... | | Authors: | Ptak, C.P, Ahmed, A.H, Oswald, R.E. | | Deposit date: | 2009-08-04 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Probing the allosteric modulator binding site of GluR2 with thiazide derivatives
Biochemistry, 48, 2009
|
|
3IJX
 
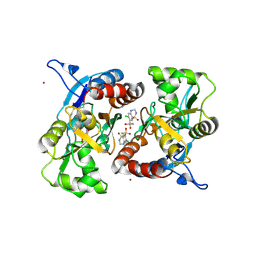 | | Crystal structure of the AMPA subunit GluR2 bound to the allosteric modulator, hydrochlorothiazide | | Descriptor: | 6-chloro-3,4-dihydro-2H-1,2,4-benzothiadiazine-7-sulfonamide 1,1-dioxide, GLUTAMIC ACID, Glutamate receptor 2, ... | | Authors: | Ptak, C.P, Ahmed, A.H, Oswald, R.E. | | Deposit date: | 2009-08-05 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.881 Å) | | Cite: | Probing the allosteric modulator binding site of GluR2 with thiazide derivatives
Biochemistry, 48, 2009
|
|
3ILU
 
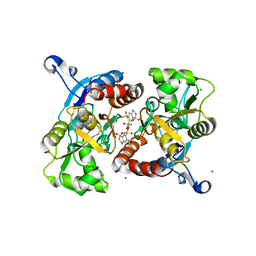 | | Crystal structure of the AMPA subunit GluR2 bound to the allosteric modulator, hydroflumethiazide | | Descriptor: | 6-(trifluoromethyl)-3,4-dihydro-2H-1,2,4-benzothiadiazine-7-sulfonamide 1,1-dioxide, GLUTAMIC ACID, Glutamate receptor 2, ... | | Authors: | Ahmed, A.H, Ptak, C.P, Oswald, R.E. | | Deposit date: | 2009-08-07 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Probing the allosteric modulator binding site of GluR2 with thiazide derivatives
Biochemistry, 48, 2009
|
|
4F29
 
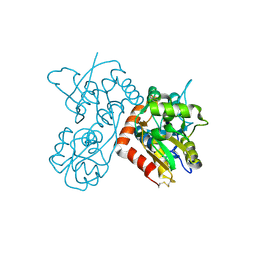 | | Quisqualate bound to the ligand binding domain of GluA3i | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Glutamate receptor 3, ZINC ION | | Authors: | Ahmed, A.H, Oswald, R.E. | | Deposit date: | 2012-05-07 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | The loss of an electrostatic contact unique to AMPA receptor ligand binding domain 2 slows channel activation.
Biochemistry, 51, 2012
|
|
4F1Y
 
 | | CNQX bound to the ligand binding domain of GluA3 | | Descriptor: | 7-nitro-2,3-dioxo-2,3-dihydroquinoxaline-6-carbonitrile, Glutamate receptor 3, ZINC ION | | Authors: | Ahmed, A.H, Oswald, R.E. | | Deposit date: | 2012-05-07 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The loss of an electrostatic contact unique to AMPA receptor ligand binding domain 2 slows channel activation.
Biochemistry, 51, 2012
|
|
4F2Q
 
 | | Quisqualate bound to the D655A mutant of the ligand binding domain of GluA3 | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Glutamate receptor 3, ZINC ION | | Authors: | Ahmed, A.H, Oswald, R.E. | | Deposit date: | 2012-05-08 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | The loss of an electrostatic contact unique to AMPA receptor ligand binding domain 2 slows channel activation.
Biochemistry, 51, 2012
|
|
4F3B
 
 | |
4F31
 
 | |
4F39
 
 | | Kainate bound to the ligand binding domain of GluA3 | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 3, ZINC ION | | Authors: | Ahmed, A.H, Oswald, R.E. | | Deposit date: | 2012-05-09 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.834 Å) | | Cite: | The loss of an electrostatic contact unique to AMPA receptor ligand binding domain 2 slows channel activation.
Biochemistry, 51, 2012
|
|
4F2O
 
 | | Quisqualate bound to the D655A mutant of the ligand binding domain of GluA3 | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Glutamate receptor 3, ZINC ION | | Authors: | Ahmed, A.H, Oswald, R.E. | | Deposit date: | 2012-05-08 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.912 Å) | | Cite: | The loss of an electrostatic contact unique to AMPA receptor ligand binding domain 2 slows channel activation.
Biochemistry, 51, 2012
|
|
4F3G
 
 | | Kainate bound to the ligand binding domain of GluA3i | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 3, ZINC ION | | Authors: | Ahmed, A.H, Oswald, R.E. | | Deposit date: | 2012-05-09 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.064 Å) | | Cite: | The loss of an electrostatic contact unique to AMPA receptor ligand binding domain 2 slows channel activation.
Biochemistry, 51, 2012
|
|
