5H04
 
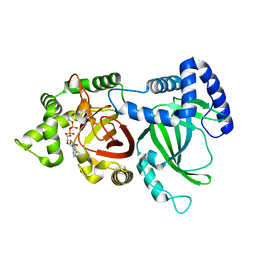 | | Crystal structure of an ADP-ribosylating toxin BECa of a novel binary enterotoxin of C. perfringens with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Binary enterotoxin of Clostridium perfringens component a | | Authors: | Kawahara, K, Yonogi, S, Munetomo, R, Oki, H, Yoshida, T, Ohkubo, T, Kumeda, Y, Matsuda, S, Kodama, T, Iida, T, Nakamura, S. | | Deposit date: | 2016-10-03 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.825 Å) | | Cite: | Crystal structure of the ADP-ribosylating component of BEC, the binary enterotoxin of Clostridium perfringens.
Biochem.Biophys.Res.Commun., 480, 2016
|
|
2HW8
 
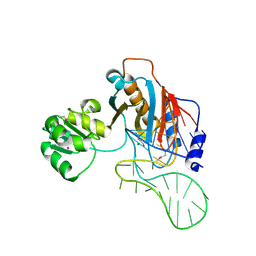 | | Structure of ribosomal protein L1-mRNA complex at 2.1 resolution. | | Descriptor: | 1,4-BUTANEDIOL, 36-MER, 50S ribosomal protein L1, ... | | Authors: | Tishchenko, S, Nikonova, E, Nikulin, A, Nevskaya, N, Volchkov, S, Piendl, W, Garber, M, Nikonov, S. | | Deposit date: | 2006-08-01 | | Release date: | 2006-12-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the ribosomal protein L1-mRNA complex at 2.1 A resolution: common features of crystal packing of L1-RNA complexes.
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
8DCP
 
 | | PI 3-kinase alpha with nanobody 3-126 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DD4
 
 | | PI 3-kinase alpha with nanobody 3-142 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DD8
 
 | | PI 3-kinase alpha with nanobody 3-142, crosslinked with DSG | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DCX
 
 | | PI 3-kinase alpha with nanobody 3-159 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4F9T
 
 | | Ribosomal protein L1 from Thermus thermophilus with substitution Thr217Ala | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 50S ribosomal protein L1, ... | | Authors: | Kljashtorny, V.G, Volchkov, S.A, Nikonova, E.Y, Kostareva, O.S, Tishchenko, S.V, Nevskaya, N.A, Nikonov, S.V. | | Deposit date: | 2012-05-21 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | [Crystal structures of mutant ribosomal proteins L1].
MOL.BIOL.(MOSCOW), 41, 2007
|
|
1SZ6
 
 | | MISTLETOE LECTIN I FROM VISCUM ALBUM. CRYSTAL STRUCTURE AT 2.05 A RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AZIDE ION, BETA-GALACTOSIDE SPECIFIC LECTIN I B CHAIN, ... | | Authors: | Gabdoulkhakov, A.G, Guhlistova, N.E, Lyashenko, A.V, Krauspenhaar, R, Stoeva, S, Voelter, W, Nikonov, S.V, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2004-04-04 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of Viscum album Mistletoe Lectin I in native state at 2.05 A resolution, comparison of structure active site conformation in ricin and in viscumin
TO BE PUBLISHED
|
|
1U1T
 
 | | Hfq protein from Pseudomonas aeruginosa. High-salt crystals | | Descriptor: | Hfq protein | | Authors: | Nikulin, A.D, Stolboushkina, E.A, Perederina, A.A, Vassilieva, I.M, Blaesi, U, Moll, I, Kachalova, G, Yokoyama, S, Vassylyev, D, Garber, M, Nikonov, S.V, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-07-16 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Pseudomonas aeruginosa Hfq protein.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1U1S
 
 | | Hfq protein from Pseudomonas aeruginosa. Low-salt crystals | | Descriptor: | Hfq protein | | Authors: | Nikulin, A.D, Stolboushkina, E.A, Perederina, I, Vassilieva, I.M, Blaesi, U, Moll, I, Kachalova, G, Vassylyev, D, Yokoyama, S, Garber, M, Nikonov, S.V, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-07-16 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Pseudomonas aeruginosa Hfq protein.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1RZO
 
 | | Agglutinin from Ricinus communis with galactoaza | | Descriptor: | Agglutinin, SULFATE ION, beta-D-galactopyranose | | Authors: | Gabdoulkhakov, A.G, Savochkina, Y, Konareva, N, Krauspenhaar, R, Stoeva, S, Nikonov, S.V, Voelter, W, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2003-12-26 | | Release date: | 2004-12-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structure-function investigation complex of Agglutinin from Ricinus communis with galactoaza
To be Published
|
|
6T8N
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K3007 | | Descriptor: | 1,2-ETHANEDIOL, Activin receptor type I, DIMETHYL SULFOXIDE, ... | | Authors: | Adamson, R.J, Williams, E.P, Bonomo, S, Rankin, S, Bacos, D, Rae, A, Cramp, S, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2019-10-24 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K3007
To Be Published
|
|
7DIH
 
 | |
1SJ9
 
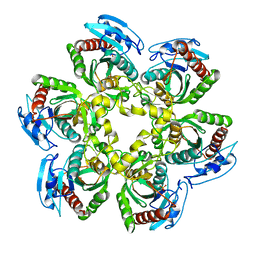 | | Crystal structure of the uridine phosphorylase from Salmonella typhimurium at 2.5A resolution | | Descriptor: | PHOSPHATE ION, Uridine phosphorylase | | Authors: | Dontsova, M, Gabdoulkhakov, A, Morgunova, E, Garber, M, Nikonov, S, Betzel, C, Ealick, S, Mikhailov, A. | | Deposit date: | 2004-03-03 | | Release date: | 2005-03-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Preliminary investigation of the three-dimensional structure of Salmonella typhimurium uridine phosphorylase in the crystalline state.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
3U4M
 
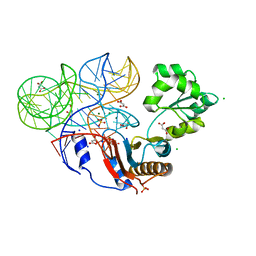 | | Crystal structure of ribosomal protein tthl1 in complex with 80nt 23s rna from thermus thermophilus | | Descriptor: | 50S ribosomal protein L1, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Gabdulkhakov, A.G, Nevskaya, N.A, NIkonov, S.V. | | Deposit date: | 2011-10-10 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution crystal structure of the isolated ribosomal L1 stalk.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4QG3
 
 | | Crystal structure of mutant ribosomal protein G219V TthL1 in complex with 80nt 23S RNA from Thermus thermophilus | | Descriptor: | 50S ribosomal protein L1, BETA-MERCAPTOETHANOL, ISOPROPYL ALCOHOL, ... | | Authors: | Gabdulkhakov, A.G, Nevskaya, N.A, Nikonov, S.V. | | Deposit date: | 2014-05-22 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein-RNA affinity of ribosomal protein L1 mutants does not correlate with the number of intermolecular interactions.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3TG8
 
 | | Mutant ribosomal protein L1 lacking ala158 from thermus thermophilus | | Descriptor: | 50S ribosomal protein L1, CHLORIDE ION, TETRAETHYLENE GLYCOL | | Authors: | Gabdulkhakov, A.G, Nevskaya, N.A, Nikonov, S.V. | | Deposit date: | 2011-08-17 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of interdomain mobility in ribosomal L1 proteins.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3U56
 
 | |
4REO
 
 | | Mutant ribosomal protein l1 from thermus thermophilus with threonine 217 replaced by valine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 50S ribosomal protein L1, GLYCINE, ... | | Authors: | Gabdulkhakov, A.G, Nevskaya, N.A, Tishchenko, S.V, Nikonov, S.V. | | Deposit date: | 2014-09-23 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Protein-RNA affinity of ribosomal protein L1 mutants does not correlate with the number of intermolecular interactions.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1RYZ
 
 | | Uridine Phosphorylase from Salmonella typhimurium. Crystal Structure at 2.9 A Resolution | | Descriptor: | ACETIC ACID, Uridine phosphorylase | | Authors: | Dontsova, M.V, Gabdoulkhakov, A.G, Lyashenko, A.V, Nikonov, S.V, Ealick, S.E, Mikhailov, A.M. | | Deposit date: | 2003-12-23 | | Release date: | 2004-12-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-functions studies of uridine phosphorylase from Salmonella typhimurium
TO BE PUBLISHED
|
|
1I6U
 
 | | RNA-PROTEIN INTERACTIONS: THE CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN S8/RRNA COMPLEX FROM METHANOCOCCUS JANNASCHII | | Descriptor: | 16S RRNA FRAGMENT, 30S RIBOSOMAL PROTEIN S8P, SULFATE ION | | Authors: | Tishchenko, S, Nikulin, A, Fomenkova, N, Nevskaya, N, Nikonov, O, Dumas, P, Moine, H, Ehresmann, B, Ehresmann, C, Piendl, W, Lamzin, V, Garber, M, Nikonov, S. | | Deposit date: | 2001-03-05 | | Release date: | 2001-08-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Detailed analysis of RNA-protein interactions within the ribosomal protein S8-rRNA complex from the archaeon Methanococcus jannaschii.
J.Mol.Biol., 311, 2001
|
|
1AN7
 
 | |
3UMY
 
 | |
3WD4
 
 | | Serratia marcescens Chitinase B complexed with azide inhibitor and quinoline compound | | Descriptor: | (E)-N-(prop-2-en-1-yloxy)-1-(quinolin-4-yl)methanimine, Chitinase B, GLYCEROL, ... | | Authors: | Hirose, T, Maita, N, Gouda, H, Koseki, J, Yamamoto, T, Sugawara, A, Nakano, H, Hirono, S, Shiomi, K, Watanabe, T, Taniguchi, H, Sharpless, K.B, Omura, S, Sunazuka, T. | | Deposit date: | 2013-06-06 | | Release date: | 2013-09-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Observation of the controlled assembly of preclick components in the in situ click chemistry generation of a chitinase inhibitor
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4QGB
 
 | | Crystal structure of mutant ribosomal protein G219V TthL1 | | Descriptor: | 50S ribosomal protein L1, ACETATE ION, CHLORIDE ION | | Authors: | Gabdulkhakov, A.G, Nevskaya, N.A, Nikonov, S.V. | | Deposit date: | 2014-05-22 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Protein-RNA affinity of ribosomal protein L1 mutants does not correlate with the number of intermolecular interactions.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
