1ZBO
 
 | | X-Ray Crystal Structure of Protein BPP1347 from Bordetella parapertussis. Northeast Structural Genomics Consortium Target BoR27. | | Descriptor: | hypothetical protein BPP1347 | | Authors: | Forouhar, F, Yong, W, Conover, K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-04-08 | | Release date: | 2005-04-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Hypothetical Protein BPP1347 from Bordetella parapertussis, Northeast Structural Genomics Target BoR27.
To be Published
|
|
3ORY
 
 | | Crystal structure of Flap endonuclease 1 from hyperthermophilic archaeon Desulfurococcus amylolyticus | | Descriptor: | PHOSPHATE ION, flap endonuclease 1 | | Authors: | Mase, T, Kubota, K, Miyazono, K, Kawarabayashii, Y, Tanokura, M. | | Deposit date: | 2010-09-08 | | Release date: | 2011-02-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of flap endonuclease 1 from the hyperthermophilic archaeon Desulfurococcus amylolyticus
Acta Crystallogr.,Sect.F, 67, 2011
|
|
1YOJ
 
 | | Crystal structure of Src kinase domain | | Descriptor: | proto-oncogene tyrosine-protein kinase SRC | | Authors: | Breitenlechner, C.B, Kairies, N.A, Honold, K, Scheiblich, S, Koll, H, Greiter, E, Koch, S, Schaefer, W, Huber, R, Engh, R.A. | | Deposit date: | 2005-01-27 | | Release date: | 2006-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of active SRC kinase domain complexes
J.Mol.Biol., 353, 2005
|
|
1YOL
 
 | | Crystal structure of Src kinase domain in complex with CGP77675 | | Descriptor: | 1-{4-[4-AMINO-5-(3-METHOXYPHENYL)-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL]BENZYL}PIPERIDIN-4-OL, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Breitenlechner, C.B, Kairies, N.A, Honold, K, Scheiblich, S, Koll, H, Greiter, E, Koch, S, Schaefer, W, Huber, R, Engh, R.A. | | Deposit date: | 2005-01-27 | | Release date: | 2006-01-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of active SRC kinase domain complexes
J.Mol.Biol., 353, 2005
|
|
1YW4
 
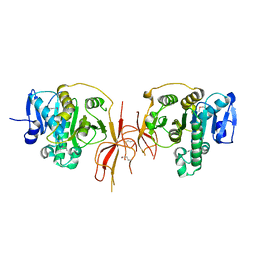 | | Crystal Structure of the Succinylglutamate Desuccinylase from Chromobacterium violaceum, Northeast Structural Genomics Target CvR22. | | Descriptor: | Succinylglutamate desuccinylase, ZINC ION | | Authors: | Forouhar, F, Abashidze, M, Conover, K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-02-17 | | Release date: | 2005-03-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Succinylglutamate Desuccinylase from Chromobacterium violaceum, Northeast Structural Genomics Target CvR22.
To be Published
|
|
3OV2
 
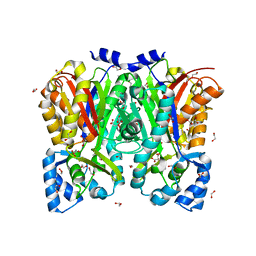 | | Curcumin synthase 1 from Curcuma longa | | Descriptor: | 1,2-ETHANEDIOL, Curcumin synthase, MALONATE ION | | Authors: | Katsuyama, Y, Miyazono, K, Tanokura, M, Ohnishi, Y, Horinouchi, S. | | Deposit date: | 2010-09-15 | | Release date: | 2010-12-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A hydrophobic cavity discovered in a curcumin synthase facilitates utilization of a beta-keto acid as an extender substrate for the atypical type III polyleteide synthase
To be Published
|
|
4JC0
 
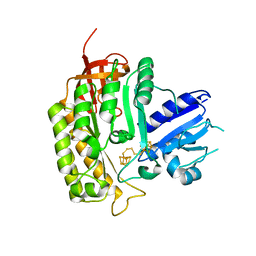 | | Crystal structure of Thermotoga maritima holo RimO in complex with pentasulfide, Northeast Structural Genomics Consortium Target VR77 | | Descriptor: | IRON/SULFUR PENTA-SULFIDE CONNECTED CLUSTERS, Ribosomal protein S12 methylthiotransferase RimO | | Authors: | Forouhar, F, Hussain, M, Seetharaman, J, Fang, Y, Chen, C.X, Cunningham, K, Conover, K, Ma, L.-C, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-02-20 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Two Fe-S clusters catalyze sulfur insertion by radical-SAM methylthiotransferases.
Nat.Chem.Biol., 9, 2013
|
|
3OV3
 
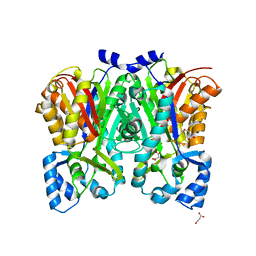 | | G211F mutant of curcumin synthase 1 from Curcuma longa | | Descriptor: | Curcumin synthase, MALONATE ION | | Authors: | Katsuyama, Y, Miyazono, K, Tanokura, M, Ohnishi, Y, Horinouchi, S. | | Deposit date: | 2010-09-15 | | Release date: | 2010-12-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A hydrophobic cavity discovered in a curcumin synthase facilitates utilization of a beta-keto acid as an extender substrate for the atypical type III polyleteide synthase
To be Published
|
|
1ZCE
 
 | | X-Ray Crystal Structure of Protein Atu2648 from Agrobacterium tumefaciens. Northeast Structural Genomics Consortium Target AtR33. | | Descriptor: | hypothetical protein Atu2648 | | Authors: | Forouhar, F, Chen, Y, Conover, K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-04-11 | | Release date: | 2005-04-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural genomics reveals EVE as a new ASCH/PUA-related domain.
Proteins, 75, 2009
|
|
3WG7
 
 | | A 1.9 angstrom radiation damage free X-ray structure of large (420KDa) protein by femtosecond crystallography | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Hirata, K, Shinzawa-Itoh, K, Yano, N, Takemura, S, Kato, K, Hatanaka, M, Muramoto, K, Kawahara, T, Tsukihara, T, Yamashita, E, Tono, K, Ueno, G, Hikima, T, Murakami, H, Inubushi, Y, Yabashi, M, Ishikawa, T, Yamamoto, M, Ogura, T, Sugimoto, H, Shen, J.R, Yoshikawa, S, Ago, H. | | Deposit date: | 2013-07-29 | | Release date: | 2014-04-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Determination of damage-free crystal structure of an X-ray-sensitive protein using an XFEL.
Nat.Methods, 11, 2014
|
|
1ZBM
 
 | | X-Ray Crystal Structure of Protein AF1704 from Archaeoglobus fulgidus. Northeast Structural Genomics Consortium Target GR62A. | | Descriptor: | hypothetical protein AF1704 | | Authors: | Forouhar, F, Abashidze, M, Conover, K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-04-08 | | Release date: | 2005-04-19 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: |
|
|
3WXQ
 
 | | Serial femtosecond X-ray structure of human fatty acid-binding protein type-3 (FABP3) in complex with stearic acid (C18:0) determined using X-ray free-electron laser at SACLA | | Descriptor: | Fatty acid-binding protein, heart, STEARIC ACID | | Authors: | Mizohata, E, Suzuki, M, Kakinouchi, K, Sugiyama, S, Murata, M, Sugahara, M, Nango, E, Tanaka, T, Tanaka, R, Tono, K, Song, C, Hatsui, T, Joti, Y, Yabashi, M, Iwata, S. | | Deposit date: | 2014-08-04 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Grease matrix as a versatile carrier of proteins for serial crystallography
Nat. Methods, 12, 2015
|
|
3VS8
 
 | | Crystal structure of type III PKS ArsC | | Descriptor: | SODIUM ION, Type III polyketide synthase | | Authors: | Satou, R, Miyanaga, A, Ozawa, H, Funa, N, Miyazono, K, Tanokura, M, Ohnishi, Y, Horinouchi, S. | | Deposit date: | 2012-04-23 | | Release date: | 2013-04-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural basis for cyclization specificity of two Azotobacter type III polyketide synthases: a single amino acid substitution reverses their cyclization specificity
J.Biol.Chem., 288, 2013
|
|
3W6V
 
 | | Crystal structure of the DNA-binding domain of AdpA, the global transcriptional factor, in complex with a target DNA | | Descriptor: | AdpA, DNA (5'-D(*AP*GP*GP*TP*TP*GP*GP*CP*GP*GP*GP*TP*TP*CP*AP*C)-3'), DNA (5'-D(*CP*TP*GP*TP*GP*AP*AP*CP*CP*CP*GP*CP*CP*AP*AP*C)-3') | | Authors: | Yao, M.D, Ohtsuka, J, Nagata, K, Miyazono, K, Ohnishi, Y, Tanokura, M. | | Deposit date: | 2013-02-22 | | Release date: | 2013-09-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Complex Structure of the DNA-binding Domain of AdpA, the Global Transcription Factor in Streptomyces griseus, and a Target Duplex DNA Reveals the Structural Basis of Its Tolerant DNA Sequence Specificity
J.Biol.Chem., 288, 2013
|
|
3W9K
 
 | | Crystal structure of thermoacidophile-specific protein STK_08120 complexed with myristic acid | | Descriptor: | FATTY ACID-BINDING PROTEIN, MYRISTIC ACID | | Authors: | Miyakawa, T, Sawano, Y, Miyazono, K, Miyauchi, Y, Hatano, K, Tanokura, M. | | Deposit date: | 2013-04-05 | | Release date: | 2013-07-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A thermoacidophile-specific protein family, DUF3211, functions as a fatty acid carrier with novel binding mode.
J.Bacteriol., 195, 2013
|
|
3VS9
 
 | | Crystal structure of type III PKS ArsC mutant | | Descriptor: | SODIUM ION, TETRAETHYLENE GLYCOL, Type III polyketide synthase | | Authors: | Satou, R, Miyanaga, A, Ozawa, H, Funa, N, Miyazono, K, Tanokura, M, Ohnishi, Y, Horinouchi, S. | | Deposit date: | 2012-04-23 | | Release date: | 2013-04-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis for cyclization specificity of two Azotobacter type III polyketide synthases: a single amino acid substitution reverses their cyclization specificity
J.Biol.Chem., 288, 2013
|
|
3AI1
 
 | | The crystal structure of L-sorbose reductase from Gluconobacter frateurii complexed with NADPH and L-sorbose reveals the structure bases of its catalytic mechanism and high substrate selectivity | | Descriptor: | NADPH-sorbose reductase | | Authors: | Kubota, K, Nagata, K, Okai, M, Miyazono, K, Tanokura, M. | | Deposit date: | 2010-05-06 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | The Crystal Structure of l-Sorbose Reductase from Gluconobacter frateurii Complexed with NADPH and l-Sorbose
J.Mol.Biol., 407, 2011
|
|
3AI3
 
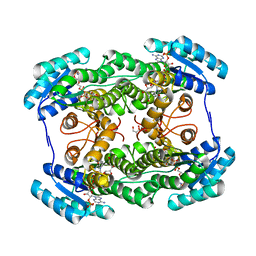 | | The crystal structure of L-Sorbose reductase from Gluconobacter frateurii complexed with NADPH and L-sorbose | | Descriptor: | L-sorbose, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH-sorbose reductase, ... | | Authors: | Kubota, K, Nagata, K, Okai, M, Miyazono, K, Tanokura, M. | | Deposit date: | 2010-05-07 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of l-Sorbose Reductase from Gluconobacter frateurii Complexed with NADPH and l-Sorbose
J.Mol.Biol., 407, 2011
|
|
3AI2
 
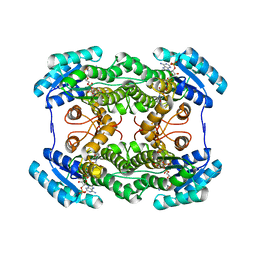 | | The crystal structure of L-sorbose reductase from Gluconobacter frateurii complexed with NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH-sorbose reductase | | Authors: | Kubota, K, Nagata, K, Okai, M, Miyazono, K, Tanokura, M. | | Deposit date: | 2010-05-07 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of l-Sorbose Reductase from Gluconobacter frateurii Complexed with NADPH and l-Sorbose
J.Mol.Biol., 407, 2011
|
|
2YT5
 
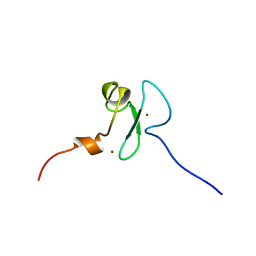 | | Solution structure of the PHD domain of Metal-response element-binding transcription factor 2 | | Descriptor: | Metal-response element-binding transcription factor 2, ZINC ION | | Authors: | Masuda, K, Muto, Y, Isono, K, Watanabe, S, Harada, T, Kigawa, T, Koseki, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PHD domain of Metal-response element-binding transcription factor 2
To be Published
|
|
2Z1N
 
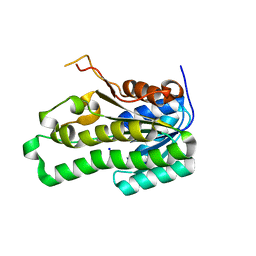 | | Crystal structure of APE0912 from Aeropyrum pernix K1 | | Descriptor: | SODIUM ION, dehydrogenase | | Authors: | Ichimura, T, Yamamura, A, Mimoto, F, Ohtsuka, J, Miyazono, K, Okai, M, Kamo, M, Lee, W.-C, Nagata, K, Tanokura, M. | | Deposit date: | 2007-05-10 | | Release date: | 2008-03-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A unique catalytic triad revealed by the crystal structure of APE0912, a short-chain dehydrogenase/reductase family protein from Aeropyrum pernix K1
Proteins, 70, 2008
|
|
3VVU
 
 | | Crystal structure of reconstructed bacterial ancestral NDK, Bac1 | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Nemoto, N, Miyazono, K, Kimura, M, Yokobori, S, Akanuma, S, Tanokura, M, Yamagishi, A. | | Deposit date: | 2012-07-27 | | Release date: | 2013-06-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Experimental evidence for the thermophilicity of ancestral life
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3VVT
 
 | | Crystal structure of reconstructed archaeal ancestral NDK, Arc1 | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Nemoto, N, Miyazono, K, Kimura, M, Yokobori, S, Akanuma, S, Tanokura, M, Yamagishi, A. | | Deposit date: | 2012-07-27 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Experimental evidence for the thermophilicity of ancestral life
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3A2E
 
 | | Crystal structure of ginkbilobin-2, the novel antifungal protein from Ginkgo biloba seeds | | Descriptor: | Ginkbilobin-2 | | Authors: | Miyakawa, T, Miyazono, K, Sawano, Y, Hatano, K, Tanokura, M. | | Deposit date: | 2009-05-13 | | Release date: | 2009-06-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of ginkbilobin-2 with homology to the extracellular domain of plant cysteine-rich receptor-like kinases
Proteins, 77, 2009
|
|
3A3K
 
 | | Reversibly bound chloride in the atrial natriuretic peptide receptor hormone-binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Atrial natriuretic peptide receptor A, ... | | Authors: | Ogawa, H, Qiu, Y, Ogata, C.M, Misono, K.S. | | Deposit date: | 2009-06-14 | | Release date: | 2010-03-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Reversibly bound chloride in the atrial natriuretic peptide receptor hormone-binding domain: Possible allosteric regulation and a conserved structural motif for the chloride-binding site.
Protein Sci., 19, 2010
|
|
