6BXM
 
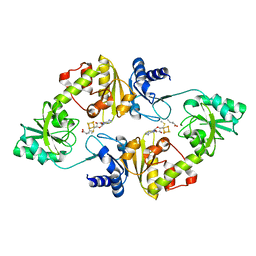 | | Crystal structure of Candidatus Methanoperedens nitroreducens Dph2 with 4Fe-4S cluster and SAM/cleaved SAM | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, ALPHA-AMINOBUTYRIC ACID, Diphthamide biosynthesis enzyme Dph2, ... | | Authors: | Fenwick, M.K, Torelli, A.T, Zhang, Y, Dong, M, Kathiresan, V, Carantoa, J.D, Dzikovski, B, Lancaster, K.M, Freed, J.H, Hoffman, B.M, Lin, H, Ealick, S.E. | | Deposit date: | 2017-12-18 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Organometallic and radical intermediates reveal mechanism of diphthamide biosynthesis.
Science, 359, 2018
|
|
6BXN
 
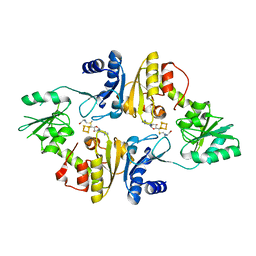 | | Crystal structure of Candidatus Methanoperedens nitroreducens Dph2 with 4Fe-4S cluster and SAM | | Descriptor: | Diphthamide biosynthesis enzyme Dph2, IRON/SULFUR CLUSTER, S-ADENOSYLMETHIONINE | | Authors: | Fenwick, M.K, Torelli, A.T, Zhang, Y, Dong, M, Kathiresan, V, Carantoa, J.D, Dzikovski, B, Lancaster, K.M, Freed, J.H, Hoffman, B.M, Lin, H, Ealick, S.E. | | Deposit date: | 2017-12-18 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.079 Å) | | Cite: | Organometallic and radical intermediates reveal mechanism of diphthamide biosynthesis.
Science, 359, 2018
|
|
5H64
 
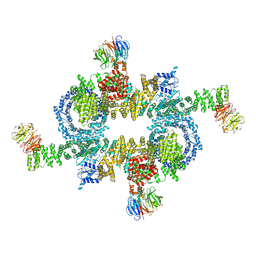 | | Cryo-EM structure of mTORC1 | | Descriptor: | Regulatory-associated protein of mTOR, Serine/threonine-protein kinase mTOR, Target of rapamycin complex subunit LST8 | | Authors: | Yang, H, Wang, J, Liu, M, Chen, X, Huang, M, Tan, D, Dong, M, Wong, C.C.L, Wang, J, Xu, Y, Wang, H. | | Deposit date: | 2016-11-10 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | 4.4 angstrom Resolution Cryo-EM structure of human mTOR Complex 1
Protein Cell, 7, 2016
|
|
5LWE
 
 | | Crystal structure of the human CC chemokine receptor type 9 (CCR9) in complex with vercirnon | | Descriptor: | C-C chemokine receptor type 9, CHOLESTEROL, MALONATE ION, ... | | Authors: | Oswald, C, Rappas, M, Kean, J, Dore, A.S, Errey, J.C, Bennett, K, Deflorian, F, Christopher, J.A, Jazayeri, A, Mason, J.S, Congreve, M, Cooke, R.M, Marshall, F.H. | | Deposit date: | 2016-09-16 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Intracellular allosteric antagonism of the CCR9 receptor.
Nature, 540, 2016
|
|
5NX2
 
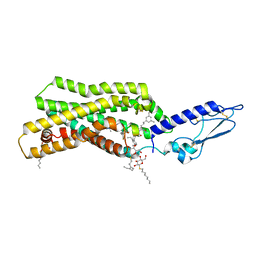 | | Crystal structure of thermostabilised full-length GLP-1R in complex with a truncated peptide agonist at 3.7 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glucagon-like peptide 1 receptor, ... | | Authors: | Rappas, M, Jazayeri, A, Brown, A.J.H, Kean, J, Errey, J.C, Robertson, N, Fiez-Vandal, C, Andrews, S.P, Congreve, M, Bortolato, A, Mason, J.S, Baig, A.H, Teobald, I, Dore, A.S, Weir, M, Cooke, R.M, Marshall, F.H. | | Deposit date: | 2017-05-09 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structure of the GLP-1 receptor bound to a peptide agonist.
Nature, 546, 2017
|
|
6BXO
 
 | | Crystal structure of Candidatus Methanoperedens nitroreducens Dph2 with 4Fe-4S cluster and SAH | | Descriptor: | Diphthamide biosynthesis enzyme Dph2, IRON/SULFUR CLUSTER, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Fenwick, M.K, Torelli, A.T, Zhang, Y, Dong, M, Kathiresan, V, Carantoa, J.D, Dzikovski, B, Lancaster, K.M, Freed, J.H, Hoffman, B.M, Lin, H, Ealick, S.E. | | Deposit date: | 2017-12-18 | | Release date: | 2018-04-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.655 Å) | | Cite: | Organometallic and radical intermediates reveal mechanism of diphthamide biosynthesis.
Science, 359, 2018
|
|
6DLN
 
 | |
6F4E
 
 | | Crystal structure of the zinc-free catalytic domain of botulinum neurotoxin X | | Descriptor: | Catalytic domain of botulinum neurotoxin X, DI(HYDROXYETHYL)ETHER | | Authors: | Masuyer, G, Henriksson, L, Kosenina, S, Zhang, S, Barkho, S, Shen, Y, Dong, M, Stenmark, P. | | Deposit date: | 2017-11-29 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural characterisation of the catalytic domain of botulinum neurotoxin X - high activity and unique substrate specificity.
Sci Rep, 8, 2018
|
|
1T49
 
 | | Allosteric Inhibition of Protein Tyrosine Phosphatase 1B | | Descriptor: | 3-(3,5-DIBROMO-4-HYDROXY-BENZOYL)-2-ETHYL-BENZOFURAN-6-SULFONIC ACID (4-SULFAMOYL-PHENYL)-AMIDE, MAGNESIUM ION, Protein-tyrosine phosphatase, ... | | Authors: | Wiesmann, C, Barr, K.J, Kung, J, Zhu, J, Shen, W, Fahr, B.J, Zhong, M, Taylor, L, Randal, M, McDowell, R.S, Hansen, S.K. | | Deposit date: | 2004-04-28 | | Release date: | 2004-07-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Allosteric inhibition of protein tyrosine phosphatase 1B.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1T4J
 
 | | Allosteric Inhibition of Protein Tyrosine Phosphatase 1B | | Descriptor: | 3-(3,5-DIBROMO-4-HYDROXY-BENZOYL)-2-ETHYL-BENZOFURAN-6-SULFONIC ACID [4-(THIAZOL-2-YLSULFAMOYL)-PHENYL]-AMIDE, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Wiesmann, C, Barr, K.J, Kung, J, Zhu, J, Shen, W, Fahr, B.J, Zhong, M, Taylor, L, Randal, M, McDowell, R.S, Hansen, S.K. | | Deposit date: | 2004-04-29 | | Release date: | 2004-07-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Allosteric inhibition of protein tyrosine phosphatase 1B
Nat.Struct.Mol.Biol., 11, 2004
|
|
5Y8Y
 
 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 1,2-ETHANEDIOL, 5-bromanyl-2-methoxy-N-(6-methoxy-3-methyl-1,2-benzoxazol-5-yl)benzenesulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Song, M, Wang, C. | | Deposit date: | 2017-08-21 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
6F47
 
 | | Crystal structure of the catalytic domain of botulinum neurotoxin X | | Descriptor: | Catalytic domain of botulinum neurotoxin X, ZINC ION | | Authors: | Masuyer, G, Henriksson, L, Kosenina, S, Zhang, S, Barkho, S, Shen, Y, Dong, M, Stenmark, P. | | Deposit date: | 2017-11-29 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural characterisation of the catalytic domain of botulinum neurotoxin X - high activity and unique substrate specificity.
Sci Rep, 8, 2018
|
|
6Q2E
 
 | | Crystal structure of Methanobrevibacter smithii Dph2 bound to 5'-methylthioadenosine | | Descriptor: | 2-(3-amino-3-carboxypropyl)histidine synthase, 5'-DEOXY-5'-METHYLTHIOADENOSINE, CHLORIDE ION, ... | | Authors: | Fenwick, M.K, Dong, M, Lin, H, Ealick, S.E. | | Deposit date: | 2019-08-07 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.768 Å) | | Cite: | The Crystal Structure of Dph2 in Complex with Elongation Factor 2 Reveals the Structural Basis for the First Step of Diphthamide Biosynthesis.
Biochemistry, 58, 2019
|
|
6Q2D
 
 | | Crystal structure of Methanobrevibacter smithii Dph2 in complex with Methanobrevibacter smithii elongation factor 2 | | Descriptor: | 2-(3-amino-3-carboxypropyl)histidine synthase, Elongation factor 2, IRON/SULFUR CLUSTER | | Authors: | Fenwick, M.K, Dong, M, Lin, H, Ealick, S.E. | | Deposit date: | 2019-08-07 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | The Crystal Structure of Dph2 in Complex with Elongation Factor 2 Reveals the Structural Basis for the First Step of Diphthamide Biosynthesis.
Biochemistry, 58, 2019
|
|
8A62
 
 | | Small molecule stabilizer (compound 2) for FOXO1 and 14-3-3 | | Descriptor: | (3~{S})-1-[3,5-bis(chloranyl)-4-oxidanyl-phenyl]carbonyl-~{N}-[2-[2-(dimethylamino)ethyldisulfanyl]ethyl]piperidine-3-carboxamide, 14-3-3 protein sigma, Forkhead box protein O1, ... | | Authors: | Kenanova, D.N, Visser, E.J, Virta, J, Sijbesma, E, Centorrino, F, Zhong, M, Vickery, H, Neitz, J, Brunsveld, L, Ottmann, C, Arkin, M.R. | | Deposit date: | 2022-06-16 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Systematic Approach to the Discovery of Protein-Protein Interaction Stabilizers.
Acs Cent.Sci., 9, 2023
|
|
8A6H
 
 | | Small molecule stabilizer (compound 7) for C-RAF and 14-3-3 | | Descriptor: | 14-3-3 protein sigma, 3-[2-(dimethylamino)ethyldisulfanyl]-1-[4-oxidanyl-4-[3-(trifluoromethyl)phenyl]piperidin-1-yl]propan-1-one, CHLORIDE ION, ... | | Authors: | Kenanova, D.N, Visser, E.J, Virta, J, Sijbesma, E, Centorrino, F, Zhong, M, Vickery, H, Neitz, J, Brunsveld, L, Ottmann, C, Arkin, M.R. | | Deposit date: | 2022-06-17 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Systematic Approach to the Discovery of Protein-Protein Interaction Stabilizers.
Acs Cent.Sci., 9, 2023
|
|
8A65
 
 | | Small molecule stabilizer (compound 3) for FOXO1 and 14-3-3 | | Descriptor: | (3~{S})-1-[2-azanyl-3,5-bis(chloranyl)phenyl]carbonyl-~{N}-[2-[2-(dimethylamino)ethyldisulfanyl]ethyl]piperidine-3-carboxamide, 14-3-3 protein sigma, Forkhead box protein O1, ... | | Authors: | Kenanova, D.N, Visser, E.J, Virta, J, Sijbesma, E, Centorrino, F, Zhong, M, Vickery, H, Neitz, J, Brunsveld, L, Ottmann, C, Arkin, M.R. | | Deposit date: | 2022-06-16 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Systematic Approach to the Discovery of Protein-Protein Interaction Stabilizers.
Acs Cent.Sci., 9, 2023
|
|
8A68
 
 | | Small molecule stabilizer (compound 5) for C-RAF(pS259) and 14-3-3 | | Descriptor: | 14-3-3 protein sigma, MAGNESIUM ION, RAF proto-oncogene serine/threonine-protein kinase, ... | | Authors: | Kenanova, D.N, Visser, E.J, Virta, J, Sijbesma, E, Centorrino, F, Zhong, M, Vickery, H, Neitz, J, Brunsveld, L, Ottmann, C, Arkin, M.R. | | Deposit date: | 2022-06-16 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Systematic Approach to the Discovery of Protein-Protein Interaction Stabilizers.
Acs Cent.Sci., 9, 2023
|
|
8A6F
 
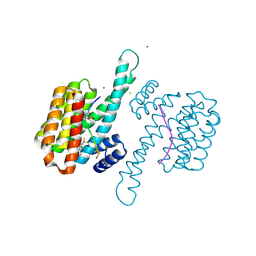 | | Small molecule stabilizer (compound 8) for C-RAF and 14-3-3 | | Descriptor: | 1-[4-[4-chloranyl-3-(trifluoromethyl)phenyl]-4-oxidanyl-piperidin-1-yl]-3-[2-(dimethylamino)ethyldisulfanyl]propan-1-one, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Kenanova, D.N, Visser, E.J, Virta, J, Sijbesma, E, Centorrino, F, Zhong, M, Vickery, H, Neitz, J, Brunsveld, L, Ottmann, C, Arkin, M.R. | | Deposit date: | 2022-06-17 | | Release date: | 2023-04-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Systematic Approach to the Discovery of Protein-Protein Interaction Stabilizers.
Acs Cent.Sci., 9, 2023
|
|
3MU3
 
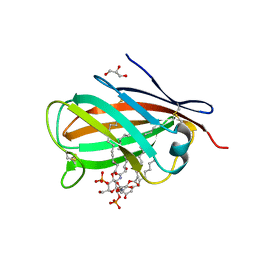 | | Crystal structure of chicken MD-1 complexed with lipid IVa | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, 2-deoxy-3-O-[(3R)-3-hydroxytetradecanoyl]-2-{[(3R)-3-hydroxytetradecanoyl]amino}-4-O-phosphono-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Yoon, S.I, Hong, M, Han, G.W, Wilson, I.A. | | Deposit date: | 2010-05-01 | | Release date: | 2010-06-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of soluble MD-1 and its interaction with lipid IVa.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6QNS
 
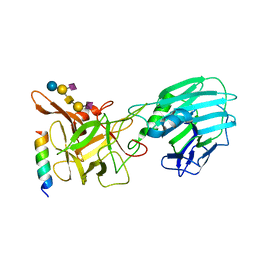 | | Crystal structure of the binding domain of Botulinum Neurotoxin type B mutant I1248W/V1249W in complex with human synaptotagmin 1 and GD1a receptors | | Descriptor: | Botulinum neurotoxin type B, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-[N-acetyl-alpha-neuraminic acid-(2-3)]beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, Synaptotagmin-1 | | Authors: | Masuyer, G, Yin, L, Zhang, S, Miyashita, S.I, Dong, M, Stenmark, P. | | Deposit date: | 2019-02-12 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Characterization of a membrane binding loop leads to engineering botulinum neurotoxin B with improved therapeutic efficacy.
Plos Biol., 18, 2020
|
|
1T48
 
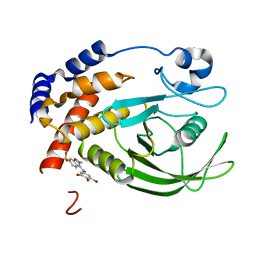 | | Allosteric Inhibition of Protein Tyrosine Phosphatase 1B | | Descriptor: | 3-(3,5-DIBROMO-4-HYDROXY-BENZOYL)-2-ETHYL-BENZOFURAN-6-SULFONIC ACID DIMETHYLAMIDE, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Wiesmann, C, Barr, K.J, Kung, J, Zhu, J, Shen, W, Fahr, B.J, Zhong, M, Erlanson, D.A, Taylor, L, Randal, M, McDowell, R.S, Hansen, S.K. | | Deposit date: | 2004-04-28 | | Release date: | 2004-07-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Allosteric inhibition of protein tyrosine phosphatase 1B
Nat.Struct.Mol.Biol., 11, 2004
|
|
3MTX
 
 | | Crystal structure of chicken MD-1 | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, GLYCEROL, Protein MD-1, ... | | Authors: | Yoon, S.I, Hong, M, Han, G.W, Wilson, I.A. | | Deposit date: | 2010-05-01 | | Release date: | 2010-06-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of soluble MD-1 and its interaction with lipid IVa.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5YFP
 
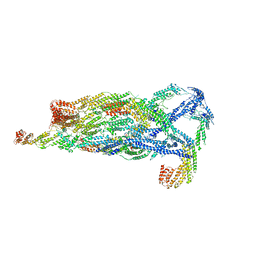 | | Cryo-EM Structure of the Exocyst Complex | | Descriptor: | Exocyst complex component EXO70, Exocyst complex component EXO84, Exocyst complex component SEC10, ... | | Authors: | Mei, K, Li, Y, Wang, S, Shao, G, Wang, J, Ding, Y, Luo, G, Yue, P, Liu, J.J, Wang, X, Dong, M.Q, Guo, W, Wang, H.W. | | Deposit date: | 2017-09-21 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structure of the exocyst complex
Nat. Struct. Mol. Biol., 25, 2018
|
|
7XM7
 
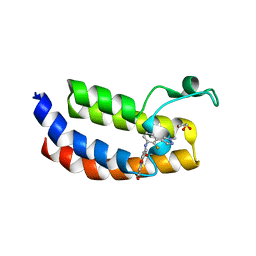 | | Crystal Structure of the CBP in complex with the Y08188 | | Descriptor: | 1,2-ETHANEDIOL, 3-ethanoyl-~{N}-[2-fluoranyl-3-(1-methylpyrazol-4-yl)phenyl]-7-methoxy-indolizine-1-carboxamide, CREB-binding protein, ... | | Authors: | Xiang, Q, Zhang, Y, Wang, C, Song, M, Xu, Y. | | Deposit date: | 2022-04-25 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Discovery and optimization of 1-(1H-indol-1-yl)ethanone derivatives as potent and selective CBP bromodomain inhibitors
To Be Published
|
|
