2J41
 
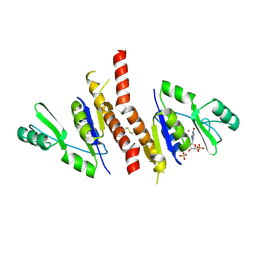 | | Crystal structure of Staphylococcus aureus guanylate monophosphate kinase | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, GUANYLATE KINASE, POTASSIUM ION, ... | | Authors: | El Omari, K, Dhaliwal, B, Lockyer, M, Charles, I, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2006-08-24 | | Release date: | 2006-10-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Staphylococcus Aureus Guanylate Monophosphate Kinase
Acta Crystallogr.,Sect.F, 62, 2006
|
|
2Y7I
 
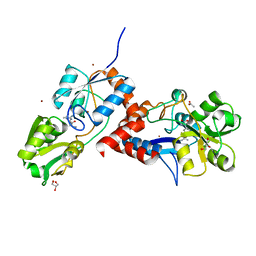 | | Structural basis for high arginine specificity in Salmonella typhimurium periplasmic binding protein STM4351. | | Descriptor: | ACETATE ION, ARGININE, GLYCEROL, ... | | Authors: | Stamp, A.L, Owen, P, El Omari, K, Lockyer, M, Lamb, H.K, Charles, I.G, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2011-01-31 | | Release date: | 2011-05-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic and Microcalorimetric Analyses Reveal the Structural Basis for High Arginine Specificity in the Salmonella Enterica Serovar Typhimurium Periplasmic Binding Protein Stm4351.
Proteins, 79, 2011
|
|
2YHM
 
 | | Structure of respiratory syncytial virus nucleocapsid protein, P212121 crystal form | | Descriptor: | NUCLEOPROTEIN, RNA | | Authors: | El Omari, K, Dhaliwal, B, Ren, J, Abrescia, N.G.A, Lockyer, M, Powell, K.L, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2011-05-04 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structures of Respiratory Syncytial Virus Nucleocapsid Protein from Two Crystal Forms: Details of Potential Packing Interactions in the Native Helical Form.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
2XJZ
 
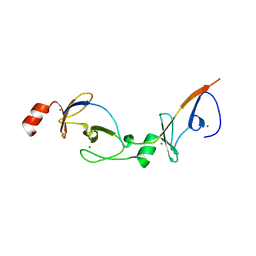 | | Crystal structure of the LMO2:LDB1-LID complex, C2 crystal form | | Descriptor: | CHLORIDE ION, LIM DOMAIN-BINDING PROTEIN 1, RHOMBOTIN-2, ... | | Authors: | El Omari, K, Karia, D, Porcher, C, Mancini, E.J. | | Deposit date: | 2010-07-06 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Leukemia Oncogene Lmo2: Implications for the Assembly of a Hematopoietic Transcription Factor Complex.
Blood, 117, 2011
|
|
2J87
 
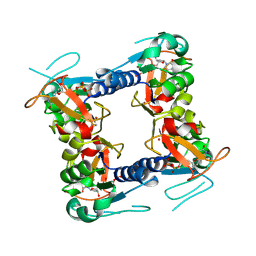 | | Structure of vaccinia virus thymidine kinase in complex with dTTP: insights for drug design | | Descriptor: | MAGNESIUM ION, THYMIDINE KINASE, THYMIDINE-5'-TRIPHOSPHATE, ... | | Authors: | El Omari, K, Solaroli, N, Karlsson, A, Balzarini, J, Stammers, D.K. | | Deposit date: | 2006-10-23 | | Release date: | 2006-11-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of Vaccinia Virus Thymidine Kinase in Complex with Dttp: Insights for Drug Design.
Bmc Struct.Biol., 6, 2006
|
|
2J0F
 
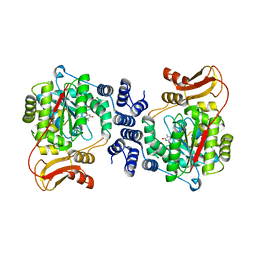 | | Structural basis for non-competitive product inhibition in human thymidine phosphorylase: implication for drug design | | Descriptor: | THYMIDINE PHOSPHORYLASE, THYMINE | | Authors: | El Omari, K, Bronckaers, A, Liekens, S, Perez-Perez, M.J, Balzarini, J, Stammers, D.K. | | Deposit date: | 2006-08-02 | | Release date: | 2006-10-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural Basis for Non-Competitive Product Inhibition in Human Thymidine Phosphorylase: Implications for Drug Design.
Biochem.J., 399, 2006
|
|
6DZ1
 
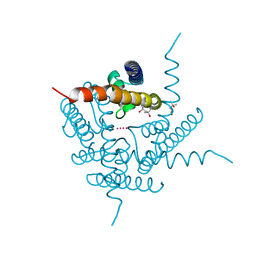 | | Studies of Ion Transport in K+ Channels | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein | | Authors: | Langan, P.S, Vandavasi, V.G, Weiss, K.L, Wagner, A, Duman, R, El Omari, K, Afonine, P.V, Coates, L. | | Deposit date: | 2018-07-02 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Anomalous X-ray diffraction studies of ion transport in K+channels.
Nat Commun, 9, 2018
|
|
7ACX
 
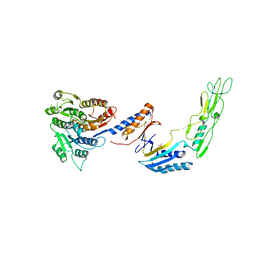 | | H/L (SLPH/SLPL) complex from C. difficile (R7404 strain) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S-layer protein, SULFATE ION | | Authors: | Lanzoni-Mangutchi, P, Barwinska-Sendra, A, Basle, A, El Omari, K, Wagner, A, Salgado, P.S. | | Deposit date: | 2020-09-11 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure and assembly of the S-layer in C. difficile.
Nat Commun, 13, 2022
|
|
3ZEU
 
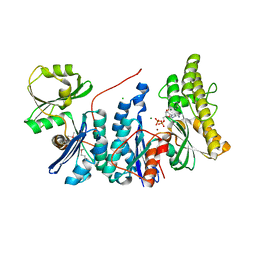 | | Structure of a Salmonella typhimurium YgjD-YeaZ heterodimer bound to ATPgammaS | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Nichols, C.E, Lamb, H.K, Thompson, P, El Omari, K, Lockyer, M, Charles, I, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-20 | | Last modified: | 2017-03-29 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Crystal Structure of the Dimer of Two Essential Salmonella Typhimurium Proteins, Ygjd & Yeaz and Calorimetric Evidence for the Formation of a Ternary Ygjd-Yeaz-Yjee Complex.
Protein Sci., 22, 2013
|
|
6QGL
 
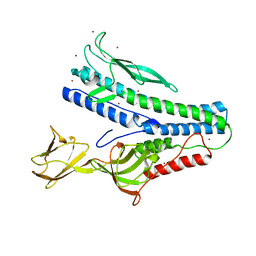 | | Crystal structure of VP5 from Haloarchaeal pleomorphic virus 6 | | Descriptor: | BROMIDE ION, VP5 | | Authors: | El Omari, K, Walter, T.S, Harlos, K, Grimes, J.M, Stuart, D.I, Roine, E. | | Deposit date: | 2019-01-11 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | The structure of a prokaryotic viral envelope protein expands the landscape of membrane fusion proteins.
Nat Commun, 10, 2019
|
|
2XF4
 
 | | Crystal structure of Salmonella enterica serovar typhimurium YcbL | | Descriptor: | HYDROXYACYLGLUTATHIONE HYDROLASE, TETRAETHYLENE GLYCOL, ZINC ION | | Authors: | Stamp, A, Owen, P, El Omari, K, Nichols, C, Lockyer, M, Lamb, H, Charles, I, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2010-05-20 | | Release date: | 2010-07-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Functional Characterization of Salmonella Enterica Serovar Typhimurium Ycbl: An Unusual Type II Glyoxalase
Protein Sci., 19, 2010
|
|
3ZET
 
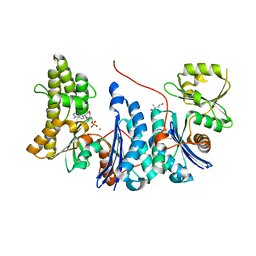 | | Structure of a Salmonella typhimurium YgjD-YeaZ heterodimer. | | Descriptor: | ADENOSINE MONOPHOSPHATE, CADMIUM ION, PROBABLE TRNA THREONYLCARBAMOYLADENOSINE BIOSYNTHESIS PROTEIN GCP, ... | | Authors: | Nichols, C.E, Lamb, H.K, Thompson, P, El Omari, K, Lockyer, M, Charles, I, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-20 | | Last modified: | 2013-05-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal Structure of the Dimer of Two Essential Salmonella Typhimurium Proteins, Ygjd & Yeaz and Calorimetric Evidence for the Formation of a Ternary Ygjd-Yeaz-Yjee Complex.
Protein Sci., 22, 2013
|
|
2YPB
 
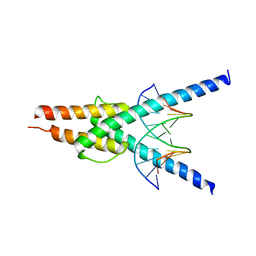 | | Structure of the SCL:E47 complex bound to DNA | | Descriptor: | EBOX FORWARD, EBOX REVERSE, T-CELL ACUTE LYMPHOCYTIC LEUKEMIA PROTEIN 1, ... | | Authors: | El Omari, K, Hoosdally, S.J, Tuladhar, K, Karia, D, Ponsele, E, Platonova, O, Vyas, P, Patient, R, Porcher, C, Mancini, E.J. | | Deposit date: | 2012-10-30 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structural Basis for Lmo2-Driven Recruitment of the Scl:E47bHLH Heterodimer to Hematopoietic-Specific Transcriptional Targets.
Cell Rep., 4, 2013
|
|
6QGI
 
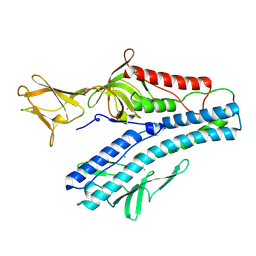 | | Crystal structure of VP5 from Haloarchaeal pleomorphic virus 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, VP5 | | Authors: | El Omari, K, Walter, T.S, Harlos, K, Grimes, J.M, Stuart, D.I, Roine, E. | | Deposit date: | 2019-01-11 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | The structure of a prokaryotic viral envelope protein expands the landscape of membrane fusion proteins.
Nat Commun, 10, 2019
|
|
2XJY
 
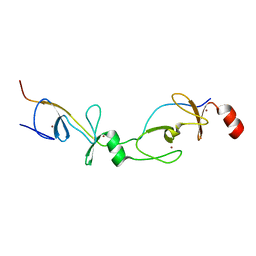 | | Crystal structure of the LMO2:LDB1-LID complex, P21 crystal form | | Descriptor: | LIM DOMAIN-BINDING PROTEIN 1, RHOMBOTIN-2, ZINC ION | | Authors: | El Omari, K, Karia, D, Porcher, C, Mancini, E.J. | | Deposit date: | 2010-07-06 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Leukemia Oncogene Lmo2: Implications for the Assembly of a Hematopoietic Transcription Factor Complex.
Blood, 117, 2011
|
|
5MUU
 
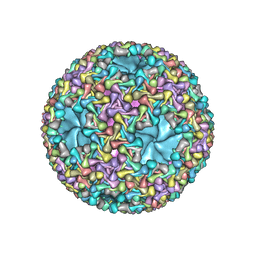 | | dsRNA bacteriophage phi6 nucleocapsid | | Descriptor: | Major inner protein P1, Major outer capsid protein, Packaging enzyme P4 | | Authors: | Sun, Z, El Omari, K, Sun, X, Ilca, S.L, Kotecha, A, Stuart, D.I, Poranen, M.M, Huiskonen, J.T. | | Deposit date: | 2017-01-14 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Double-stranded RNA virus outer shell assembly by bona fide domain-swapping.
Nat Commun, 8, 2017
|
|
7QEA
 
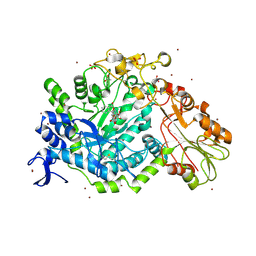 | | Crystal structure of fluorescein-di-Beta-D-glucuronide bound to a mutant of SN243 (D415A) | | Descriptor: | (2~{S},3~{S},4~{S},5~{R},6~{S})-3,4,5-tris(oxidanyl)-6-[(1~{R})-6'-oxidanyl-3-oxidanylidene-spiro[2-benzofuran-1,9'-xanthene]-3'-yl]oxy-oxane-2-carboxylic acid, ACETATE ION, SN243, ... | | Authors: | Neun, S, Brear, P, Campbell, E, Omari, K, Wagner, O, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2021-12-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Functional metagenomic screening identifies an unexpected beta-glucuronidase.
Nat.Chem.Biol., 18, 2022
|
|
7QG4
 
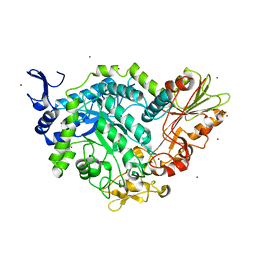 | | Apo crystal structure of a mutant of SN243 (D415N) | | Descriptor: | SN243, SULFATE ION, ZINC ION | | Authors: | Neun, S, Brear, P, Campbell, E, Omari, K, Wagner, O, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2021-12-07 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Functional metagenomic screening identifies an unexpected beta-glucuronidase.
Nat.Chem.Biol., 18, 2022
|
|
7QEF
 
 | | Crystal structure of para-nitrophenyl-Beta-D-glucuronide bound to a mutant of SN243 (D415A) | | Descriptor: | 4-nitrophenyl beta-D-glucopyranosiduronic acid, ACETATE ION, SN243, ... | | Authors: | Neun, S, Brear, P, Campbell, E, Omari, K, Wagner, O, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2021-12-02 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Functional metagenomic screening identifies an unexpected beta-glucuronidase.
Nat.Chem.Biol., 18, 2022
|
|
7QE1
 
 | | Crystal structure of apo SN243 | | Descriptor: | SN243, ZINC ION | | Authors: | Neun, S, Brear, P, Campbell, E, Omari, K, Wagner, O, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2021-12-01 | | Release date: | 2022-10-12 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Functional metagenomic screening identifies an unexpected beta-glucuronidase.
Nat.Chem.Biol., 18, 2022
|
|
7QE2
 
 | | Crystal structure of D-glucuronic acid bound to SN243 | | Descriptor: | ACETATE ION, SN243, SULFATE ION, ... | | Authors: | Neun, S, Brear, P, Campbell, E, Omari, K, Wagner, O, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2021-12-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Functional metagenomic screening identifies an unexpected beta-glucuronidase.
Nat.Chem.Biol., 18, 2022
|
|
7QEE
 
 | | SN243 mutant D415N bound to para-nitrophenyl-Beta-D-glucuronide | | Descriptor: | 4-nitrophenyl beta-D-glucopyranosiduronic acid, SN243, SULFATE ION, ... | | Authors: | Neun, S, Brear, P, Campbell, E, Omari, K, Wagner, O, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2021-12-02 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.374 Å) | | Cite: | Functional metagenomic screening identifies an unexpected beta-glucuronidase.
Nat.Chem.Biol., 18, 2022
|
|
5E8C
 
 | | pseudorabies virus nuclear egress complex, pUL31, pUL34 | | Descriptor: | CHLORIDE ION, UL31, UL34 protein, ... | | Authors: | Zeev-Ben-Mordehai, T, Cheleski, J, Whittle, C, El Omari, K, Harlos, K, Hagen, C, Klupp, B, Mettenleiter, T.C, Gruenewald, K. | | Deposit date: | 2015-10-14 | | Release date: | 2015-12-23 | | Last modified: | 2016-01-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Herpesvirus Nuclear Egress Complex Provides Insights into Inner Nuclear Membrane Remodeling.
Cell Rep, 13, 2015
|
|
6OWX
 
 | | Spy H96L:Im7 L18pI-Phe complex; multiple anomalous datasets contained herein for element identification | | Descriptor: | CHLORIDE ION, IMIDAZOLE, IODIDE ION, ... | | Authors: | Rocchio, S, Duman, R, El Omari, K, Mykhaylyk, V, Yan, Z, Wagner, A, Bardwell, J.C.A, Horowitz, S. | | Deposit date: | 2019-05-12 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Identifying dynamic, partially occupied residues using anomalous scattering.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6OWY
 
 | | Spy H96L:Im7 K20pI-Phe complex; multiple anomalous datasets contained herein for element identification | | Descriptor: | CHLORIDE ION, IMIDAZOLE, IODIDE ION, ... | | Authors: | Rocchio, S, Duman, R, El Omari, K, Mykhaylyk, V, Yan, Z, Wagner, A, Bardwell, J.C.A, Horowitz, S. | | Deposit date: | 2019-05-12 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Identifying dynamic, partially occupied residues using anomalous scattering.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
