1HOT
 
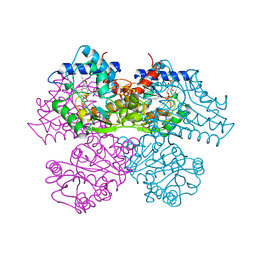 | | GLUCOSAMINE 6-PHOSPHATE DEAMINASE COMPLEXED WITH THE ALLOSTERIC ACTIVATOR N-ACETYL-GLUCOSAMINE-6-PHOSPHATE | | Descriptor: | 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, GLUCOSAMINE 6-PHOSPHATE DEAMINASE, PHOSPHATE ION | | Authors: | Oliva, G, Fontes, M.L, Garratt, R, Altamirano, M.M, Calcagno, M.L, Horjales, E. | | Deposit date: | 1995-11-17 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and catalytic mechanism of glucosamine 6-phosphate deaminase from Escherichia coli at 2.1 A resolution.
Structure, 3, 1995
|
|
6MH3
 
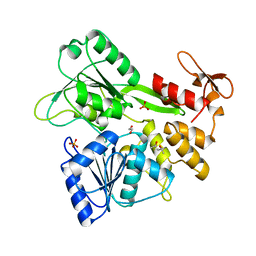 | | The crystal structure of Zika virus NS3 helicase domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, PHOSPHATE ION, ... | | Authors: | Oliva, G, Mesquita, N, Godoy, A. | | Deposit date: | 2018-09-17 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The crystal structure of Zika virus NS3 helicase domain
To Be Published
|
|
1DEA
 
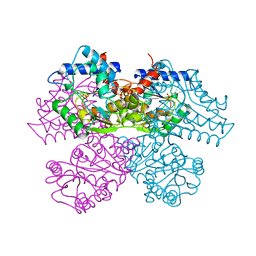 | | STRUCTURE AND CATALYTIC MECHANISM OF GLUCOSAMINE 6-PHOSPHATE DEAMINASE FROM ESCHERICHIA COLI AT 2.1 ANGSTROMS RESOLUTION | | Descriptor: | GLUCOSAMINE 6-PHOSPHATE DEAMINASE, PHOSPHATE ION | | Authors: | Oliva, G, Fontes, M.R.M, Garratt, R.C, Altamirano, M.M, Calcagno, M.L, Horjales, E. | | Deposit date: | 1995-09-13 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and catalytic mechanism of glucosamine 6-phosphate deaminase from Escherichia coli at 2.1 A resolution.
Structure, 3, 1995
|
|
1HOR
 
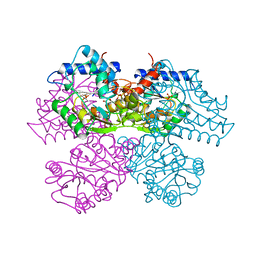 | | STRUCTURE AND CATALYTIC MECHANISM OF GLUCOSAMINE 6-PHOSPHATE DEAMINASE FROM ESCHERICHIA COLI AT 2.1 ANGSTROMS RESOLUTION | | Descriptor: | 2-DEOXY-2-AMINO GLUCITOL-6-PHOSPHATE, GLUCOSAMINE 6-PHOSPHATE DEAMINASE, PHOSPHATE ION | | Authors: | Oliva, G, Fontes, M.R.M, Garratt, R.C, Altamirano, M.M, Calcagno, M.L, Horjales, E. | | Deposit date: | 1995-09-13 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and catalytic mechanism of glucosamine 6-phosphate deaminase from Escherichia coli at 2.1 A resolution.
Structure, 3, 1995
|
|
3K1E
 
 | | Crystal structure of odorant binding protein 1 (AaegOBP1) from Aedes aegypti | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Leite, N.R, Krogh, R, Leal, W.S, Iulek, J, Oliva, G. | | Deposit date: | 2009-09-27 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of an odorant-binding protein from the mosquito Aedes aegypti suggests a binding pocket covered by a pH-sensitive "Lid".
Plos One, 4, 2009
|
|
1QXS
 
 | | CRYSTAL STRUCTURE OF Trypanosoma cruzi GLYCERALDEHYDE-3- PHOSPHATE DEHYDROGENASE COMPLEXED WITH AN ANALOGUE OF 1,3- BisPHOSPHO-D-GLYCERIC ACID | | Descriptor: | 3-HYDROXY-2-OXO-4-PHOPHONOXY- BUTYL)-PHOSPHONIC ACID, Glyceraldehyde 3-phosphate dehydrogenase, glycosomal, ... | | Authors: | Castilho, M.S, Pavao, F, Oliva, G. | | Deposit date: | 2003-09-08 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of Trypanosoma cruzi glyceraldehyde-3-phosphate dehydrogenase complexed with an analogue of 1,3-bisphospho-d-glyceric acid.
Eur.J.Biochem., 270, 2003
|
|
3OIS
 
 | | Crystal Structure Xylellain, a cysteine protease from Xylella fastidiosa | | Descriptor: | Cysteine protease, URIDINE-5'-DIPHOSPHATE | | Authors: | Leite, N.R, Faro, A.R, Oliva, M.A.V, Thiemann, O.H, Oliva, G. | | Deposit date: | 2010-08-19 | | Release date: | 2011-08-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure of the cysteine protease Xylellain from Xylella fastidiosa reveals an intriguing activation mechanism.
Febs Lett., 587, 2013
|
|
7MBG
 
 | | SARS-CoV-2 Main protease in orthorhombic space group | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Douangamath, A, von Delft, F, Noske, G.D, Nakamura, A.M, Gawriljuk, V.O, Lima, G.M.A, Zeri, A.C.M, Nascimento, A.F.Z, Oliva, G, Godoy, A.S. | | Deposit date: | 2021-03-31 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | A Crystallographic Snapshot of SARS-CoV-2 Main Protease Maturation Process.
J.Mol.Biol., 433, 2021
|
|
7ME0
 
 | | Cryo-EM structure of SARS-CoV-2 NSP15 NendoU at pH 6.0 | | Descriptor: | Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Song, Y, Nakamura, A.M, Noske, G.D, Gawriljuk, V.O, Fernandes, R.S, Oliva, G. | | Deposit date: | 2021-04-06 | | Release date: | 2021-04-14 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
7RB0
 
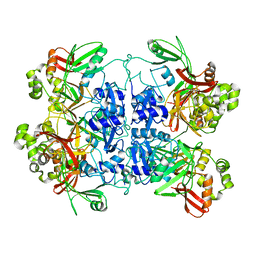 | | Cryo-EM structure of SARS-CoV-2 NSP15 NendoU at pH 7.5 | | Descriptor: | Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Song, Y, Nakamura, A.M, Noske, G.D, Gawriljuk, V.O, Fernandes, R.S, Oliva, G. | | Deposit date: | 2021-07-05 | | Release date: | 2021-07-14 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 51, 2023
|
|
7RB2
 
 | | Cryo-EM structure of SARS-CoV-2 NSP15 NendoU in BIS-Tris pH 6.0 | | Descriptor: | Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Song, Y, Nakamura, A.M, Noske, G.D, Gawriljuk, V.O, Fernandes, R.S, Oliva, G. | | Deposit date: | 2021-07-05 | | Release date: | 2021-07-14 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 51, 2023
|
|
7S82
 
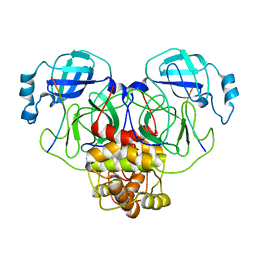 | |
6URV
 
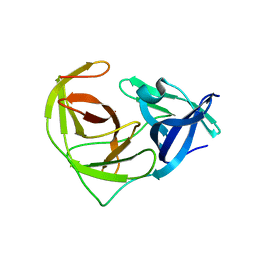 | | Crystal structure of Yellow Fever Virus NS2B-NS3 protease domain | | Descriptor: | NS2B, NS3 protease | | Authors: | Noske, G.D, Gawriljuk, V.F.O, Fernandes, R.S, Oliva, G, Godoy, A.S. | | Deposit date: | 2019-10-24 | | Release date: | 2020-01-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural characterization and polymorphism analysis of the NS2B-NS3 protease from the 2017 Brazilian circulating strain of Yellow Fever virus.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
8EYJ
 
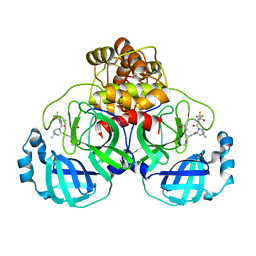 | | Crystal Structure of uncleaved SARS-CoV-2 Main Protease C145S mutant in complex with Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Noske, G.D, Godoy, A.S, Oliva, G. | | Deposit date: | 2022-10-27 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.738 Å) | | Cite: | An in-solution snapshot of SARS-COV-2 main protease maturation process and inhibition.
Nat Commun, 14, 2023
|
|
8E26
 
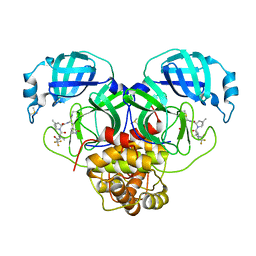 | | Crystal Structure of SARS-CoV-2 Main Protease N142S mutant in complex with Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Noske, G.D, Godoy, A.S, Oliva, G. | | Deposit date: | 2022-08-14 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease.
J.Biol.Chem., 299, 2023
|
|
8E25
 
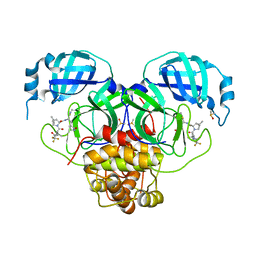 | | Crystal Structure of SARS-CoV-2 Main Protease M49I mutant in complex with Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, DIMETHYL SULFOXIDE, Replicase polyprotein 1ab | | Authors: | Noske, G.D, Godoy, A.S, Oliva, G. | | Deposit date: | 2022-08-14 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.868 Å) | | Cite: | Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease.
J.Biol.Chem., 299, 2023
|
|
1SAC
 
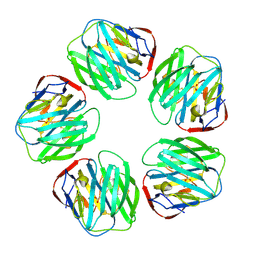 | | THE STRUCTURE OF PENTAMERIC HUMAN SERUM AMYLOID P COMPONENT | | Descriptor: | ACETIC ACID, CALCIUM ION, SERUM AMYLOID P COMPONENT | | Authors: | White, H.E, Emsley, J, O'Hara, B.P, Oliva, G, Srinivasan, N, Tickle, I.J, Blundell, T.L, Pepys, M.B, Wood, S.P. | | Deposit date: | 1994-01-27 | | Release date: | 1994-05-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of pentameric human serum amyloid P component.
Nature, 367, 1994
|
|
1CD5
 
 | | GLUCOSAMINE-6-PHOSPHATE DEAMINASE FROM E.COLI, T CONFORMER | | Descriptor: | PROTEIN (GLUCOSAMINE 6-PHOSPHATE DEAMINASE) | | Authors: | Horjales, E, Altamirano, M.M, Calcagno, M.L, Garratt, R.C, Oliva, G. | | Deposit date: | 1999-03-05 | | Release date: | 2000-03-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The allosteric transition of glucosamine-6-phosphate deaminase: the structure of the T state at 2.3 A resolution.
Structure Fold.Des., 7, 1999
|
|
8DH6
 
 | | Cryo-EM structure of Saccharomyces cerevisiae cytochrome c oxidase (Complex IV) extracted in lipid nanodiscs | | Descriptor: | CALCIUM ION, COPPER (II) ION, Cytochrome c oxidase subunit 1, ... | | Authors: | Godoy, A.S, Song, Y, Cheruvara, H, Quigley, A, Oliva, G. | | Deposit date: | 2022-06-25 | | Release date: | 2022-07-20 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Cryo-EM structure of Saccharomyces cerevisiae cytochrome c oxidase (Complex IV) extracted in lipid nanodiscs
To Be Published
|
|
5U04
 
 | | Crystal structure of Zika virus NS5 RNA-dependent RNA polymerase | | Descriptor: | PHOSPHATE ION, ZINC ION, Zika Virus NS5 RdRp | | Authors: | Godoy, A.S, Lima, G.M.A, Oliveira, K.I.Z, Torres, N.U, Maluf, F.V, Guido, R.V.C, Oliva, G. | | Deposit date: | 2016-11-22 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Zika virus NS5 RNA-dependent RNA polymerase.
Nat Commun, 8, 2017
|
|
3IDS
 
 | | Structure of Glycosomal Glyceraldehyde-3-Phosphate Dehydrogenase from Trypanosoma cruzi in complex with the irreversible iodoacetamide inhibitor | | Descriptor: | ACETAMIDE, GLYCEROL, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Balliano, T.L, Guido, R.V.C, Andricopulo, A.D, Oliva, G. | | Deposit date: | 2009-07-21 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Glycosomal Glyceraldehyde-3-Phosphate Dehydrogenase from Trypanosoma cruzi in complex with the irreversible iodoacetamide inhibitor
To be Published
|
|
3IEX
 
 | | Schistosoma Purine nucleoside phosphorylase in complex with guanosine | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, GUANOSINE, ... | | Authors: | Castilho, M.S, Pereira, H.M, Oliva, G, Andricopulo, A.D. | | Deposit date: | 2009-07-23 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for selective inhibition of purine nucleoside phosphorylase from Schistosoma mansoni: kinetic and structural studies
Bioorg.Med.Chem., 18, 2010
|
|
8DH7
 
 | |
8UM3
 
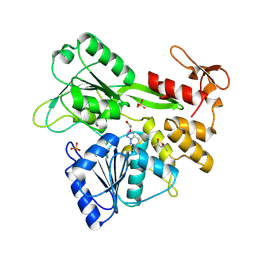 | | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z203039992 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 6-chlorotetrazolo[1,5-b]pyridazine, ... | | Authors: | Godoy, A.S, Noske, G.D, Fairhead, M, Lithgo, R.M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Mesquita, N.C.M.R, Oliva, G, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-10-17 | | Release date: | 2023-11-01 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.925 Å) | | Cite: | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z203039992
To Be Published
|
|
8V7R
 
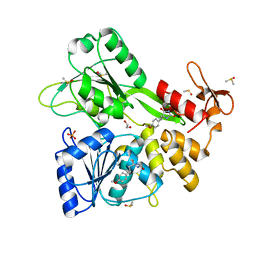 | | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z56772132 | | Descriptor: | (5R)-5-[2-(4-methoxyphenyl)ethyl]-5-methylimidazolidine-2,4-dione, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Godoy, A.S, Noske, G.D, Fairhead, M, Lithgo, R.M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Mesquita, N.C.M.R, Oliva, G, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-12-04 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z56772132
To Be Published
|
|
