1ML3
 
 | | Evidences for a flip-flop catalytic mechanism of Trypanosoma cruzi glyceraldehyde-3-phosphate dehydrogenase, from its crystal structure in complex with reacted irreversible inhibitor 2-(2-phosphono-ethyl)-acrylic acid 4-nitro-phenyl ester | | Descriptor: | (3-FORMYL-BUT-3-ENYL)-PHOSPHONIC ACID, Glyceraldehyde 3-phosphate dehydrogenase, glycosomal, ... | | Authors: | Castilho, M.S, Pavao, F, Oliva, G. | | Deposit date: | 2002-08-29 | | Release date: | 2003-07-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evidence for the Two Phosphate Binding Sites of an Analogue of the Thioacyl Intermediate for the Trypanosoma cruzi Glyceraldehyde-3-phosphate Dehydrogenase-Catalyzed Reaction, from Its Crystal Structure.
Biochemistry, 42, 2003
|
|
1MZV
 
 | | Crystal Structure of Adenine Phosphoribosyltransferase (APRT) From Leishmania tarentolae | | Descriptor: | ADENOSINE MONOPHOSPHATE, Adenine Phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Thiemann, O.H, Silva, M, Oliva, G, Silva, C.H.T.P, Iulek, J. | | Deposit date: | 2002-10-10 | | Release date: | 2003-10-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of adenine phosphoribosyltransferase from Leishmania tarentolae: potential implications for APRT catalytic mechanism.
Biochim.Biophys.Acta, 1696, 2004
|
|
1II2
 
 | | Crystal Structure of Phosphoenolpyruvate Carboxykinase (PEPCK) from Trypanosoma cruzi | | Descriptor: | PHOSPHOENOLPYRUVATE CARBOXYKINASE, SULFATE ION | | Authors: | Trapani, S, Linss, J, Goldenberg, S, Fischer, H, Craievich, A.F, Oliva, G. | | Deposit date: | 2001-04-20 | | Release date: | 2001-11-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the dimeric phosphoenolpyruvate carboxykinase (PEPCK) from Trypanosoma cruzi at 2 A resolution.
J.Mol.Biol., 313, 2001
|
|
3E9R
 
 | | Crystal structure of purine nucleoside phosphorylase from Schistosoma mansoni in complex with adenine | | Descriptor: | ACETATE ION, ADENINE, DIMETHYL SULFOXIDE, ... | | Authors: | Pereira, H.M, Rezende, M.M, Oliva, G, Garratt, R.C. | | Deposit date: | 2008-08-23 | | Release date: | 2009-09-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Adenosine binding to low-molecular-weight purine nucleoside phosphorylase: the structural basis for recognition based on its complex with the enzyme from Schistosoma mansoni.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3FB1
 
 | |
3DJF
 
 | | Crystal Structure of Schistosoma mansoni Purine Nucleoside Phosphorylase in a complex with BCX-34 | | Descriptor: | 2-amino-7-(pyridin-3-ylmethyl)-3,5-dihydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, Purine-nucleoside phosphorylase, ... | | Authors: | Postigo, M.P, Pereira, H.M, Oliva, G, Andricopulo, A.D. | | Deposit date: | 2008-06-23 | | Release date: | 2009-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Structural basis for selective inhibition of purine nucleoside phosphorylase from Schistosoma mansoni: kinetic and structural studies.
Bioorg.Med.Chem., 18, 2010
|
|
3FAZ
 
 | | Crystal structure of Schistosoma mansoni purine nucleoside phosphorylase in complex with inosine | | Descriptor: | INOSINE, Purine-nucleoside phosphorylase, SULFATE ION | | Authors: | Pereira, H.M, Garratt, R.C, Oliva, G. | | Deposit date: | 2008-11-18 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Adenosine binding to low-molecular-weight purine nucleoside phosphorylase: the structural basis for recognition based on its complex with the enzyme from Schistosoma mansoni.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3DMT
 
 | | Structure of Glycosomal Glyceraldehyde-3-Phosphate Dehydrogenase from Trypanosoma cruzi in complex with the irreversible iodoacetate inhibitor | | Descriptor: | GLYCEROL, Glyceraldehyde-3-phosphate dehydrogenase, glycosomal, ... | | Authors: | Guido, R.V.C, Balliano, T.L, Andricopulo, A.D, Oliva, G. | | Deposit date: | 2008-07-01 | | Release date: | 2008-10-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Kinetic and Crystallographic Studies on Glyceraldehyde-3-Phosphate Dehydrogenase from Trypanosoma cruzi in Complex with Iodoacetate.
Letters in drug design & discovery, 6, 2009
|
|
3F8W
 
 | | Crystal structure of Schistosoma mansoni purine nucleoside phosphorylase in complex with adenosine | | Descriptor: | ADENOSINE, DIMETHYL SULFOXIDE, Purine-nucleoside phosphorylase, ... | | Authors: | Pereira, H.M, Rezende, M.M, Garratt, R.C, Oliva, G. | | Deposit date: | 2008-11-13 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Adenosine binding to low-molecular-weight purine nucleoside phosphorylase: the structural basis for recognition based on its complex with the enzyme from Schistosoma mansoni.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3E9Z
 
 | | Crystal structure of purine nucleoside phosphorylase from Schistosoma mansoni in complex with 6-chloroguanine | | Descriptor: | 6-chloroguanine, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Pereira, H.M, Rezende, M.M, Oliva, G, Garratt, R.C. | | Deposit date: | 2008-08-24 | | Release date: | 2009-09-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal structure of Schistosoma mansoni purine nucleoside phosphorylase (SmPNP) in complex with adenine, 8-aminoguanine, 8-azaguanine and 6-chloroguanine.
To be Published
|
|
3FNQ
 
 | | Crystal structure of schistosoma purine nucleoside phosphorylase in complex with hypoxanthine | | Descriptor: | DIMETHYL SULFOXIDE, HYPOXANTHINE, Purine-nucleoside phosphorylase, ... | | Authors: | Castilho, M.S, Pereira, H.M, Garratt, R.C, Oliva, G. | | Deposit date: | 2008-12-26 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Adenosine binding to low-molecular-weight purine nucleoside phosphorylase: the structural basis for recognition based on its complex with the enzyme from Schistosoma mansoni.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3DSL
 
 | | The Three-dimensional Structure of Bothropasin, the Main Hemorrhagic Factor from Bothrops jararaca venom. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, FUROYL-LEUCINE, ... | | Authors: | Muniz, J.R.C, Ambrosio, A, Selistre-de-Araujo, H.S, Oliva, G, Garratt, R.C, Souza, D.H.F. | | Deposit date: | 2008-07-13 | | Release date: | 2008-10-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The three-dimensional structure of bothropasin, the main hemorrhagic factor from Bothrops jararaca venom: Insights for a new classification of snake venom metalloprotease subgroups.
Toxicon, 52, 2008
|
|
3FP5
 
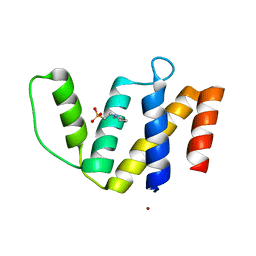 | | Crystal structure of ACBP from Moniliophthora perniciosa | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Acyl-CoA Binding Protein, ZINC ION | | Authors: | Monzani, P.S, Pereira, H.M, Melo, F.A, Meirelles, F.V, Oliva, G, Cascardo, J.C.M. | | Deposit date: | 2009-01-04 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | A new topology of ACBP from Moniliophthora perniciosa.
Biochim.Biophys.Acta, 1804, 2010
|
|
5S72
 
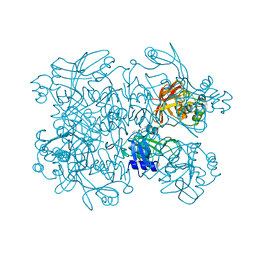 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with BBL029427 | | Descriptor: | CITRIC ACID, N-(2-aminoethyl)-N'-phenylurea, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2020-11-13 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.512 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5S6Y
 
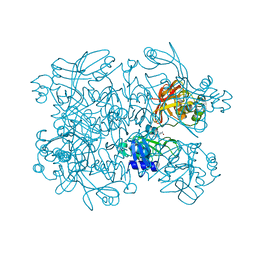 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z56900771 | | Descriptor: | CITRIC ACID, N-[(furan-2-yl)methyl]urea, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2020-11-13 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5S6Z
 
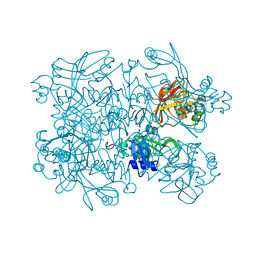 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with PB2255187532 | | Descriptor: | 4-[(dimethylamino)methyl]-1,3-thiazol-2-amine, CITRIC ACID, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2020-11-13 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5S71
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with FUZS-5 | | Descriptor: | 5'-thiothymidine, CITRIC ACID, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Bege, M, Bajusz, D, BorbAs, A, Keseru, G.M, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2020-11-13 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.941 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5S6X
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z2889976755 | | Descriptor: | 1-(2,4-dimethyl-1H-imidazol-5-yl)methanamine, CITRIC ACID, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2020-11-13 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5S70
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with EN300-181428 | | Descriptor: | (5R)-2-methyl-4,5,6,7-tetrahydro-1H-benzimidazol-5-amine, CITRIC ACID, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2020-11-13 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.327 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5SAF
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with EN300-321461 | | Descriptor: | 6,7-dihydro-5H-pyrrolo[2,3-d]pyrimidine, CITRIC ACID, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5SAC
 
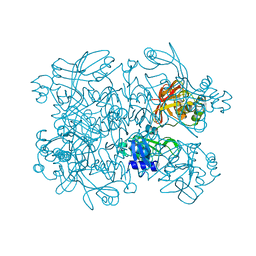 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z59181945 | | Descriptor: | CITRIC ACID, N-hydroxyquinoline-2-carboxamide, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.029 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5SA5
 
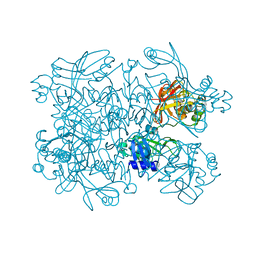 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z1530301542 | | Descriptor: | 4-ethyl-2-(1H-imidazol-5-yl)-1,3-thiazole, CITRIC ACID, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5SAA
 
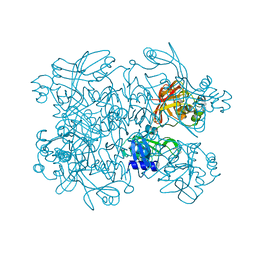 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z319891284 | | Descriptor: | 3-[(2S)-1-(methanesulfonyl)pyrrolidin-2-yl]-5-methyl-1,2-oxazole, CITRIC ACID, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.239 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5SAB
 
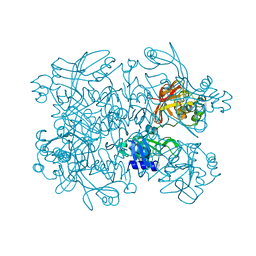 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z31504642 | | Descriptor: | 2-methoxy-N-phenylacetamide, CITRIC ACID, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.486 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
5SAE
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NendoU in complex with Z3219959731 | | Descriptor: | CITRIC ACID, Uridylate-specific endoribonuclease, pyridazin-3(2H)-one | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
