5SZZ
 
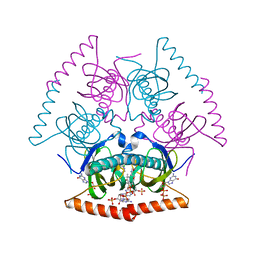 | | Novel Structural Insights into GDP-Mediated Regulation of Acyl-CoA Thioesterases | | Descriptor: | Acyl-CoA hydrolase, CHLORIDE ION, COENZYME A, ... | | Authors: | Khandokar, Y.B, Srivastava, P, Forwood, J.K. | | Deposit date: | 2016-08-15 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into GDP-mediated regulation of a bacterial acyl-CoA thioesterase.
J. Biol. Chem., 292, 2017
|
|
5EGJ
 
 | |
5HWF
 
 | |
4I82
 
 | |
4XY6
 
 | |
3W6I
 
 | | Crystal structure of 19F probe-labeled hCAI | | Descriptor: | 1-(2-ethoxyethoxy)-3,5-bis(trifluoromethyl)benzene, Carbonic anhydrase 1, ZINC ION | | Authors: | Takaoka, Y, Kioi, Y, Morito, A, Otani, J, Arita, K, Ashihara, E, Ariyoshi, M, Tochio, H, Shirakawa, M, Hamachi, I. | | Deposit date: | 2013-02-14 | | Release date: | 2013-03-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Quantitative Comparison of Protein Dynamics in Live Cells and In Vitro by In-Cell 19F-NMR
To be published
|
|
4IEN
 
 | | Crystal Structure of Acyl-CoA Hydrolase from Neisseria meningitidis FAM18 | | Descriptor: | CHLORIDE ION, COENZYME A, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Khandokar, Y.B, Forwood, J.K. | | Deposit date: | 2012-12-13 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Expression, purification and crystallization of acetyl-CoA hydrolase from Neisseria meningitidis.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4P1W
 
 | |
4P1N
 
 | | Crystal structure of Atg1-Atg13 complex | | Descriptor: | Atg1 tMIT, Atg13 MIM | | Authors: | Fujioka, Y, Noda, N.N. | | Deposit date: | 2014-02-27 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of starvation-induced assembly of the autophagy initiation complex.
Nat.Struct.Mol.Biol., 21, 2014
|
|
3W6H
 
 | | Crystal structure of 19F probe-labeled hCAI in complex with acetazolamide | | Descriptor: | 1-(2-ethoxyethoxy)-3,5-bis(trifluoromethyl)benzene, 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, Carbonic anhydrase 1, ... | | Authors: | Takaoka, Y, Kioi, Y, Morito, A, Otani, J, Arita, K, Ashihara, E, Ariyoshi, M, Tochio, H, Shirakawa, M, Hamachi, I. | | Deposit date: | 2013-02-14 | | Release date: | 2013-03-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.964 Å) | | Cite: | Quantitative Comparison of Protein Dynamics in Live Cells and In Vitro by In-Cell 19F-NMR
To be published
|
|
5AXE
 
 | | Crystal Structure Analysis of DNA Duplexes containing sulfoamide-bridged nucleic acid (SuNA-NH) | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(LSH)P*AP*CP*GP*C)-3') | | Authors: | Mitsuoka, Y, Aoyama, H, Kugimiya, A, Fujimura, Y, Yamamoto, T, Waki, R, Wada, F, Tahara, S, Sawamura, M, Noda, M, Hari, Y, Obika, S. | | Deposit date: | 2015-07-28 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Effect of an N-substituent in sulfonamide-bridged nucleic acid (SuNA) on hybridization ability and duplex structure.
Org.Biomol.Chem., 14, 2016
|
|
1JDA
 
 | | MALTOTETRAOSE-FORMING EXO-AMYLASE | | Descriptor: | 1,4-ALPHA MALTOTETRAHYDROLASE, CALCIUM ION | | Authors: | Yoshioka, Y, Hasegawa, K, Matsuura, Y, Katsube, Y, Kubota, M. | | Deposit date: | 1997-06-16 | | Release date: | 1997-10-15 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of a mutant maltotetraose-forming exo-amylase cocrystallized with maltopentaose.
J.Mol.Biol., 271, 1997
|
|
1QPK
 
 | | MUTANT (D193G) MALTOTETRAOSE-FORMING EXO-AMYLASE IN COMPLEX WITH MALTOTETRAOSE | | Descriptor: | CALCIUM ION, PROTEIN (MALTOTETRAOSE-FORMING AMYLASE), alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yoshioka, Y, Hasegawa, K, Matsuura, Y, Katsube, Y, Kubota, M. | | Deposit date: | 1999-05-26 | | Release date: | 1999-11-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Roles of catalytic residues in alpha-amylases as evidenced by the structures of the product-complexed mutants of a maltotetraose-forming amylase.
Protein Eng., 12, 1999
|
|
5AXF
 
 | | Crystal Structure Analysis of DNA Duplexes containing sulfoamide-bridged nucleic acid (SuNA-NMe) | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(LSM)P*AP*CP*GP*C)-3') | | Authors: | Mitsuoka, Y, Aoyama, H, Kugimiya, A, Fujimura, Y, Yamamoto, T, Waki, R, Wada, F, Tahara, S, Sawamura, M, Noda, M, Hari, Y, Obika, S. | | Deposit date: | 2015-07-28 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Effect of an N-substituent in sulfonamide-bridged nucleic acid (SuNA) on hybridization ability and duplex structure.
Org.Biomol.Chem., 14, 2016
|
|
3WHN
 
 | | Hemerythrin-like domain of DcrH I119H mutant (met) | | Descriptor: | CALCIUM ION, CHLORO DIIRON-OXO MOIETY, Hemerythrin-like domain protein DcrH | | Authors: | Okamoto, Y, Onoda, A, Sugimoto, H, Takano, Y, Hirota, S, Kurtz Jr, D.M, Shiro, Y, Hayashi, T. | | Deposit date: | 2013-08-29 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | H2O2-dependent substrate oxidation by an engineered diiron site in a bacterial hemerythrin.
Chem.Commun.(Camb.), 50, 2014
|
|
3WAQ
 
 | | Hemerythrin-like domain of DcrH I119E mutant (met) | | Descriptor: | Hemerythrin-like domain protein DcrH, MU-OXO-DIIRON | | Authors: | Okamoto, Y, Onoda, A, Sugimoto, H, Takano, Y, Hirota, S, Kurtz Jr, D.M, Shiro, Y, Hayashi, T. | | Deposit date: | 2013-05-07 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure, exogenous ligand binding, and redox properties of an engineered diiron active site in a bacterial hemerythrin
Inorg.Chem., 52, 2013
|
|
3A7P
 
 | |
3A7O
 
 | |
1BMQ
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF INTERLEUKIN-1BETA CONVERTING ENZYME (ICE) WITH A PEPTIDE BASED INHIBITOR, (3S )-N-METHANESULFONYL-3-({1-[N-(2-NAPHTOYL)-L-VALYL]-L-PROLYL }AMINO)-4-OXOBUTANAMIDE | | Descriptor: | (3S)-N-METHANESULFONYL-3-({1-[N-(2-NAPHTOYL)-L-VALYL]-L-PROLYL}AMINO)-4-OXOBUTANAMIDE, PROTEIN (INTERLEUKIN-1 BETA CONVERTASE) | | Authors: | Okamoto, Y, Anan, H, Nakai, E, Morihira, K, Yonetoku, Y, Kurihara, H, Katayama, N, Sakashita, H, Terai, Y, Takeuchi, M, Shibanuma, T, Isomura, Y. | | Deposit date: | 1998-07-24 | | Release date: | 1998-07-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Peptide based interleukin-1 beta converting enzyme (ICE) inhibitors: synthesis, structure activity relationships and crystallographic study of the ICE-inhibitor complex.
Chem.Pharm.Bull., 47, 1999
|
|
8JUD
 
 | | Crystal structure of human MMP-7 in complex with inhibitor | | Descriptor: | CALCIUM ION, Matrilysin, Peptide Inhibitor, ... | | Authors: | Kamitani, M, Abe-Sato, K, Oka, Y. | | Deposit date: | 2023-06-26 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Based Optimization and Biological Evaluation of Potent and Selective MMP-7 Inhibitors for Kidney Fibrosis.
J.Med.Chem., 66, 2023
|
|
8JUF
 
 | | Crystal structure of human MMP-7 in complex with inhibitor | | Descriptor: | CALCIUM ION, Matrilysin, Peptide Inhibitor, ... | | Authors: | Kamitani, M, Abe-Sato, K, Oka, Y. | | Deposit date: | 2023-06-26 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structure-Based Optimization and Biological Evaluation of Potent and Selective MMP-7 Inhibitors for Kidney Fibrosis.
J.Med.Chem., 66, 2023
|
|
8JUG
 
 | | Crystal structure of human MMP-7 in complex with inhibitor | | Descriptor: | CALCIUM ION, Matrilysin, Peptide Inhibitor, ... | | Authors: | Kamitani, M, Abe-Sato, K, Oka, Y. | | Deposit date: | 2023-06-26 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-Based Optimization and Biological Evaluation of Potent and Selective MMP-7 Inhibitors for Kidney Fibrosis.
J.Med.Chem., 66, 2023
|
|
8K4Z
 
 | |
7WXX
 
 | | Crystal structure of human MMP-7 in complex with inhibitor | | Descriptor: | CALCIUM ION, Matrilysin, Peptide Inhibitor, ... | | Authors: | Kamitani, M, Oka, Y, Tabuse, H. | | Deposit date: | 2022-02-15 | | Release date: | 2022-10-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of Highly Potent and Selective Matrix Metalloproteinase-7 Inhibitors by Hybridizing the S1' Subsite Binder with Short Peptides.
J.Med.Chem., 65, 2022
|
|
6AJE
 
 | | Crystal structure of DHODH in complex with ferulenol from Eimeria tenella | | Descriptor: | 4-oxidanyl-3-[(2~{E},6~{E})-3,7,11-trimethyldodeca-2,6,10-trienyl]chromen-2-one, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Shiba, T, Inaoka, D.K, Sato, D, Hartuti, E.D, Amalia, E, Nagahama, M, Yoshioka, Y, Matsubayashi, M, Balogun, E.O, Tsuji, N, Kita, K, Harada, S. | | Deposit date: | 2018-08-27 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Structural and Biochemical Features of Eimeria tenella Dihydroorotate Dehydrogenase, a Potential Drug Target.
Genes (Basel), 11, 2020
|
|
