6H96
 
 | | AlbA-albicidin complex, albicidin resistance protein | | Descriptor: | 4-[[4-[[4-[(3~{S})-5-azanyl-3-[[4-[[(~{E})-3-(4-hydroxyphenyl)-2-methyl-prop-2-enoyl]amino]phenyl]carbonylamino]-2-oxidanylidene-3~{H}-pyrrol-1-yl]phenyl]carbonylamino]-3-methoxy-2-oxidanyl-phenyl]carbonylamino]-3-methoxy-2-oxidanyl-benzoic acid, Albicidin resistance protein, SULFATE ION | | Authors: | Koehnke, J, Sikandar, A. | | Deposit date: | 2018-08-03 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Adaptation of a Bacterial Multidrug Resistance System Revealed by the Structure and Function of AlbA.
J.Am.Chem.Soc., 140, 2018
|
|
6H95
 
 | | AlbA, albicidin resistance protein | | Descriptor: | Albicidin resistance protein | | Authors: | Koehnke, J, Sikandar, A. | | Deposit date: | 2018-08-03 | | Release date: | 2018-11-21 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Adaptation of a Bacterial Multidrug Resistance System Revealed by the Structure and Function of AlbA.
J.Am.Chem.Soc., 140, 2018
|
|
6HAI
 
 | | AlbAM131A mutant in complex with albicidin , albicidin resistance protein | | Descriptor: | 4-[[4-[[4-[(3~{S})-3-[[4-[[(~{E})-3-(4-hydroxyphenyl)-2-methyl-prop-2-enoyl]amino]phenyl]carbonylamino]-2,5-bis(oxidanylidene)pyrrolidin-1-yl]phenyl]carbonylamino]-3-methoxy-2-oxidanyl-phenyl]carbonylamino]-3-methoxy-2-oxidanyl-benzoic acid, Albicidin resistance protein, SULFATE ION | | Authors: | Koehnke, J, Sikandar, A. | | Deposit date: | 2018-08-07 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Adaptation of a Bacterial Multidrug Resistance System Revealed by the Structure and Function of AlbA.
J.Am.Chem.Soc., 140, 2018
|
|
3BOD
 
 | | Structure of mouse beta-neurexin 1 | | Descriptor: | CALCIUM ION, Neurexin-1-alpha | | Authors: | Koehnke, J, Jin, X, Shapiro, L. | | Deposit date: | 2007-12-17 | | Release date: | 2008-03-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of beta-Neurexin 1 and beta-Neurexin 2 Ectodomains and Dynamics of Splice Insertion Sequence 4.
Structure, 16, 2008
|
|
3BOP
 
 | | Structure of mouse beta-neurexin 2D4 | | Descriptor: | beta-Neurexin 2D4 | | Authors: | Koehnke, J, Jin, X, Shapiro, L. | | Deposit date: | 2007-12-17 | | Release date: | 2008-03-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structures of beta-Neurexin 1 and beta-Neurexin 2 Ectodomains and Dynamics of Splice Insertion Sequence 4.
Structure, 16, 2008
|
|
8CIO
 
 | |
5O28
 
 | | E. coli FolD apo | | Descriptor: | Bifunctional protein FolD | | Authors: | Koehnke, J, Sikandar, A. | | Deposit date: | 2017-05-19 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The natural product carolacton inhibits folate-dependent C1 metabolism by targeting FolD/MTHFD.
Nat Commun, 8, 2017
|
|
6H97
 
 | | AlbAT99V mutant , albicidin resistance protein | | Descriptor: | Albicidin resistance protein | | Authors: | Koehnke, J, Sikandar, A. | | Deposit date: | 2018-08-03 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Adaptation of a Bacterial Multidrug Resistance System Revealed by the Structure and Function of AlbA.
J.Am.Chem.Soc., 140, 2018
|
|
5OFX
 
 | | Plla lectin, trisaccharide complex | | Descriptor: | CALCIUM ION, PllA, alpha-D-galactopyranose-(1-6)-alpha-D-glucopyranose-(1-2)-beta-D-fructofuranose | | Authors: | Koehnke, J, Sikandar, A. | | Deposit date: | 2017-07-11 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Photorhabdus luminescens lectin A (PllA): A new probe for detecting alpha-galactoside-terminating glycoconjugates.
J. Biol. Chem., 292, 2017
|
|
5OFZ
 
 | | PllA lectin, apo | | Descriptor: | Uncharacterized protein | | Authors: | Koehnke, J, Sikandar, A. | | Deposit date: | 2017-07-11 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Photorhabdus luminescens lectin A (PllA): A new probe for detecting alpha-galactoside-terminating glycoconjugates.
J. Biol. Chem., 292, 2017
|
|
5ODU
 
 | | PllA lectin, monosaccharide complex | | Descriptor: | CALCIUM ION, PllA, methyl alpha-D-galactopyranoside | | Authors: | Koehnke, J, Sikandar, A. | | Deposit date: | 2017-07-06 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Photorhabdus luminescens lectin A (PllA): A new probe for detecting alpha-galactoside-terminating glycoconjugates.
J. Biol. Chem., 292, 2017
|
|
4B0Z
 
 | | Crystal structure of S. pombe Rpn12 | | Descriptor: | 26S PROTEASOME REGULATORY SUBUNIT RPN12, GLYCEROL, MONOTHIOGLYCEROL, ... | | Authors: | Boehringer, J, Riedinger, C, Paraskevopoulos, K, Johnson, E.O.D, Lowe, E.D, Khoudian, C, Smith, D, Noble, M.E.M, Gordon, C, Endicott, J.A. | | Deposit date: | 2012-07-06 | | Release date: | 2012-09-12 | | Last modified: | 2012-11-07 | | Method: | X-RAY DIFFRACTION (1.585 Å) | | Cite: | Structural and Functional Characterisation of Rpn12 Identifies Residues Required for Rpn10 Proteasome Incorporation.
Biochem.J., 448, 2012
|
|
5OFI
 
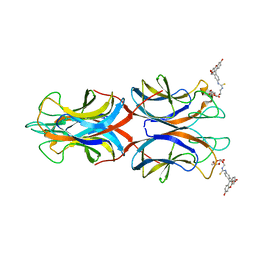 | | PllA lectin, Fluorophore carbohydrate complex | | Descriptor: | 5-[2-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxyethylcarbamothioylamino]-2-(6-oxidanyl-3-oxidanylidene-4,9-dihydroxanthen-9-yl)benzoic acid, CALCIUM ION, PIIA | | Authors: | Koehnke, J, Sikandar, A. | | Deposit date: | 2017-07-11 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Photorhabdus luminescens lectin A (PllA): A new probe for detecting alpha-galactoside-terminating glycoconjugates.
J. Biol. Chem., 292, 2017
|
|
3ZXX
 
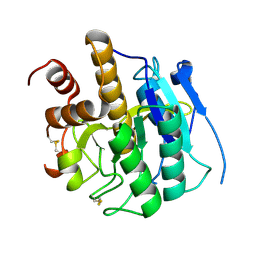 | | Structure of self-cleaved protease domain of PatA | | Descriptor: | SUBTILISIN-LIKE PROTEIN | | Authors: | Koehnke, J, Zollman, D, Vendome, J, Raab, A, Houssen, W.E, Smith, M.C, Jaspars, M, Naismith, J.H. | | Deposit date: | 2011-08-16 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Discovery of New Cyanobactins from Cyanothece Pcc 7425 Defines a New Signature for Processing of Patellamides.
Chembiochem, 13, 2012
|
|
4AKS
 
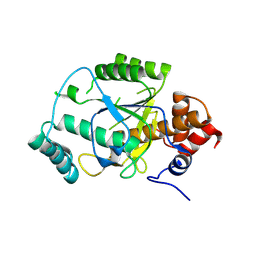 | | PatG macrocyclase domain | | Descriptor: | THIAZOLINE OXIDASE/SUBTILISIN-LIKE PROTEASE | | Authors: | Koehnke, J, Bent, A, Houssen, W.E, Zollman, D, Morawitz, F, Shirran, S, Vendome, J, Nneoyiegbe, A.F, Trembleau, L, Botting, C.H, Smith, M.C.M, Jaspars, M, Naismith, J.H. | | Deposit date: | 2012-02-28 | | Release date: | 2012-07-18 | | Last modified: | 2013-11-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The Mechanism of Patellamide Macrocyclization Revealed by the Characterization of the Patg Macrocyclase Domain.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3ZXY
 
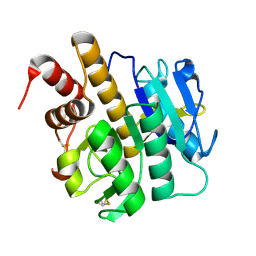 | | Structure of S218A mutant of the protease domain of PatA | | Descriptor: | SUBTILISIN-LIKE PROTEIN | | Authors: | Koehnke, J, Zollman, D, Vendome, J, Raab, A, Houssen, W.E, Smith, M.C, Jaspars, M, Naismith, J.H. | | Deposit date: | 2011-08-16 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The Discovery of New Cyanobactins from Cyanothece Pcc 7425 Defines a New Signature for Processing of Patellamides.
Chembiochem, 13, 2012
|
|
4AKT
 
 | | PatG macrocyclase in complex with peptide | | Descriptor: | SUBSTRATE ANALOGUE, THIAZOLINE OXIDASE/SUBTILISIN-LIKE PROTEASE | | Authors: | Koehnke, J, Bent, A, Houssen, W.E, Zollman, D, Morawitz, F, Shirran, S, Vendome, J, Nneoyiegbe, A.F, Trembleau, L, Botting, C.H, Smith, M.C.M, Jaspars, M, Naismith, J.H. | | Deposit date: | 2012-02-28 | | Release date: | 2012-07-18 | | Last modified: | 2013-11-06 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | The Mechanism of Patellamide Macrocyclization Revealed by the Characterization of the Patg Macrocyclase Domain.
Nat.Struct.Mol.Biol., 19, 2012
|
|
6YF6
 
 | |
6ZSV
 
 | | Structure of crocagin biosynthetic protein CgnB | | Descriptor: | Uncharacterized protein, ZINC ION | | Authors: | Koehnke, J, Adam, S. | | Deposit date: | 2020-07-16 | | Release date: | 2022-07-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Unusual peptide-binding proteins guide pyrroloindoline alkaloid formation in crocagin biosynthesis.
Nat.Chem., 15, 2023
|
|
6ZYL
 
 | | non-heme monooxygenase; ThoJ apo | | Descriptor: | L(+)-TARTARIC ACID, Uncharacterized protein | | Authors: | Koehnke, J, Sikandar, A. | | Deposit date: | 2020-08-02 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Non-Heme Monooxygenase ThoJ Catalyzes Thioholgamide beta-Hydroxylation.
Acs Chem.Biol., 15, 2020
|
|
6ZYK
 
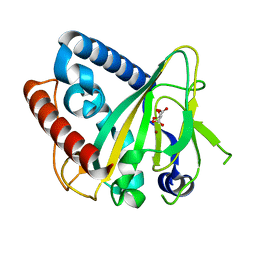 | | Non-heme monooxygenase, ThoJ-Ni complex | | Descriptor: | L(+)-TARTARIC ACID, Monooxygenase, NICKEL (II) ION | | Authors: | Koehnke, J, Sikandar, A. | | Deposit date: | 2020-08-02 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Non-Heme Monooxygenase ThoJ Catalyzes Thioholgamide beta-Hydroxylation.
Acs Chem.Biol., 15, 2020
|
|
4BS9
 
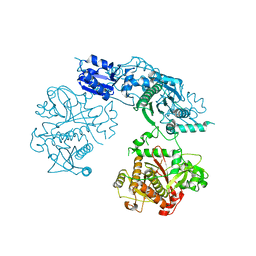 | | Structure of the heterocyclase TruD | | Descriptor: | TRUD, ZINC ION | | Authors: | Koehnke, J, Bent, A.F, Zollman, D, Smith, K, Houssen, W.E, Zhu, X, Mann, G, Lebl, T, Scharff, R, Shirran, S, Botting, C.H, Jaspars, M, Schwarz-Linek, U, Naismith, J.H. | | Deposit date: | 2013-06-09 | | Release date: | 2013-11-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Cyanobactin Heterocyclase Enzyme: A Processive Adenylase that Operates with a Defined Order of Reaction.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
4LVA
 
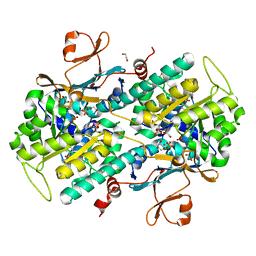 | | Fragment-based Identification of Amides Derived From trans-2-(Pyridin-3-yl)cyclopropanecarboxylic Acid as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | 1,2-ETHANEDIOL, N-(4-{[4-(pyrrolidin-1-yl)piperidin-1-yl]sulfonyl}benzyl)-2H-pyrido[4,3-e][1,2,4]thiadiazin-3-amine 1,1-dioxide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Giannetti, A.M, Zheng, X, Skelton, N, Wang, W, Bravo, B, Feng, Y, Gunzner-Toste, J, Ho, Y, Hua, R, Wang, C, Zhao, Q, Liederer, B.M, Liu, Y, O'Brien, T, Oeh, J, Sampath, D, Shen, Y, Wang, L, Wu, H, Xiao, Y, Yuen, P, Zak, M, Zhao, G, Dragovich, P.S. | | Deposit date: | 2013-07-26 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification of amides derived from 1H-pyrazolo[3,4-b]pyridine-5-carboxylic acid as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4LVG
 
 | | Fragment-based Identification of Amides Derived From trans-2-(Pyridin-3-yl)cyclopropanecarboxylic Acid as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | (1S,2S)-N-[4-(phenylsulfonyl)phenyl]-2-(pyridin-3-yl)cyclopropanecarboxamide, 1,2-ETHANEDIOL, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Giannetti, A.M, Zheng, X, Skelton, N, Wang, W, Bravo, B, Feng, Y, Gunzner-Toste, J, Ho, Y, Hua, R, Wang, C, Zhao, Q, Liederer, B.M, Liu, Y, O'Brien, T, Oeh, J, Sampath, D, Shen, Y, Wang, L, Wu, H, Xiao, Y, Yuen, P, Zak, M, Zhao, G, Dragovich, P.S. | | Deposit date: | 2013-07-26 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Identification of amides derived from 1H-pyrazolo[3,4-b]pyridine-5-carboxylic acid as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4M6P
 
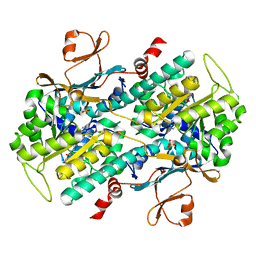 | | Identification of Amides Derived From 1H-Pyrazolo[3,4-b]pyridine-5-carboxylic Acid as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | N-[4-(phenylsulfonyl)benzyl]-2H-pyrazolo[3,4-b]pyridine-5-carboxamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Giannetti, A.M, Zheng, X, Skelton, N, Wang, W, Bravo, B, Feng, Y, Gunzner-Toste, J, Ho, Y, Hua, R, Wang, C, Zhao, Q, Liederer, B.M, Liu, Y, O'Brien, T, Oeh, J, Sampath, D, Shen, Y, Wang, L, Wu, H, Xiao, Y, Yuen, P, Zak, M, Zhao, G, Dragovich, P.S. | | Deposit date: | 2013-08-10 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification of amides derived from 1H-pyrazolo[3,4-b]pyridine-5-carboxylic acid as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
