5I13
 
 | | Endonuclease inhibitor 2 bound to influenza strain H1N1 polymerase acidic subunit N-terminal region at pH 7.0 | | Descriptor: | 4-{(E)-[2-(4-chlorophenyl)hydrazinylidene]methyl}benzene-1,2,3-triol, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Fudo, S, Yamamoto, N, Nukaga, M, Odagiri, T, Tashiro, M, Hoshino, T. | | Deposit date: | 2016-02-05 | | Release date: | 2016-02-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Two Distinctive Binding Modes of Endonuclease Inhibitors to the N-Terminal Region of Influenza Virus Polymerase Acidic Subunit
Biochemistry, 55, 2016
|
|
6EHM
 
 | | Model of the Ebola virus nucleocapsid subunit from recombinant virus-like particles | | Descriptor: | Membrane-associated protein VP24, Nucleoprotein | | Authors: | Wan, W, Kolesnikova, L, Clarke, M, Koehler, A, Noda, T, Becker, S, Briggs, J.A.G. | | Deposit date: | 2017-09-13 | | Release date: | 2017-11-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Structure and assembly of the Ebola virus nucleocapsid.
Nature, 551, 2017
|
|
5YQ0
 
 | | Crystal structure of secreted protein CofJ from ETEC. | | Descriptor: | CALCIUM ION, CofJ | | Authors: | Oki, H, Kawahara, K, Maruno, T, Imai, T, Muroga, Y, Fukakusa, S, Iwashita, T, Kobayashi, Y, Matsuda, S, Kodama, T, Iida, T, Yoshida, T, Ohkubo, T, Nakamura, S. | | Deposit date: | 2017-11-04 | | Release date: | 2018-06-27 | | Last modified: | 2018-07-25 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Interplay of a secreted protein with type IVb pilus for efficient enterotoxigenicEscherichia colicolonization
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YPZ
 
 | | Crystal structure of minor pilin CofB from CFA/III complexed with N-terminal peptide fragment of CofJ | | Descriptor: | CofB, CofJ | | Authors: | Oki, H, Kawahara, K, Maruno, T, Imai, T, Muroga, Y, Fukakusa, S, Iwashita, T, Kobayashi, Y, Matsuda, S, Kodama, T, Iida, T, Yoshida, T, Ohkubo, T, Nakamura, S. | | Deposit date: | 2017-11-04 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.521 Å) | | Cite: | Interplay of a secreted protein with type IVb pilus for efficient enterotoxigenicEscherichia colicolonization.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7Z20
 
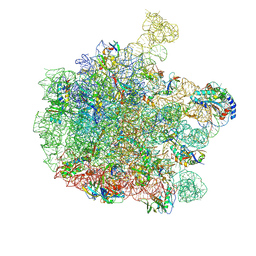 | | 70S E. coli ribosome with an extended uL23 loop from Candidatus marinimicrobia and a stalled filamin domain 5 nascent chain | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Mitropoulou, A, Plessa, E, Wlodarski, T, Ahn, M, Sidhu, H, Becker, T.A, Beckmann, R, Cabrita, L.D, Christodoulou, J. | | Deposit date: | 2022-02-25 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | Modulating co-translational protein folding by rational design and ribosome engineering.
Nat Commun, 13, 2022
|
|
7ZP8
 
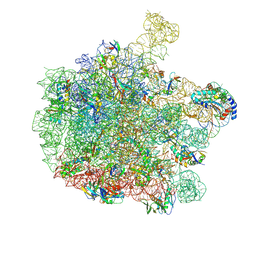 | | 70S E. coli ribosome with a stalled filamin domain 5 nascent chain | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Mitropoulou, A, Plessa, E, Wlodarski, T, Ahn, M, Chan, S.H.S, Becker, T.A, Beckmann, R, Cabrita, L.D, Christodoulou, J. | | Deposit date: | 2022-04-26 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Modulating co-translational protein folding by rational design and ribosome engineering.
Nat Commun, 13, 2022
|
|
7ZQ5
 
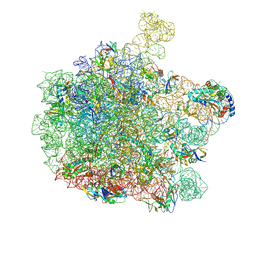 | | 70S E. coli ribosome with truncated uL23 and uL24 loops | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Mitropoulou, A, Wlodarski, T, Ahn, M, Becker, T.A, Beckmann, R, Cabrita, L.D, Christodoulou, J. | | Deposit date: | 2022-04-29 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Modulating co-translational protein folding by rational design and ribosome engineering.
Nat Commun, 13, 2022
|
|
7ZQ6
 
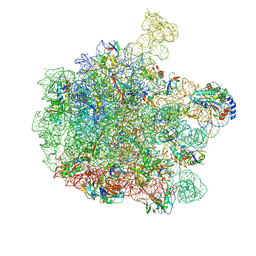 | | 70S E. coli ribosome with truncated uL23 and uL24 loops and a stalled filamin domain 5 nascent chain | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Mitropoulou, A, Wlodarski, T, Ahn, M, Becker, T.A, Beckmann, R, Cabrita, L.D, Christodoulou, J. | | Deposit date: | 2022-04-29 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Modulating co-translational protein folding by rational design and ribosome engineering.
Nat Commun, 13, 2022
|
|
7ZOD
 
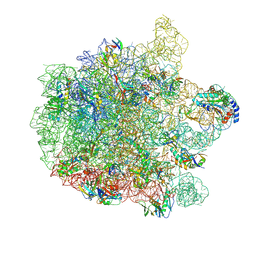 | | 70S E. coli ribosome with an extended uL23 loop from Candidatus marinimicrobia | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Mitropoulou, A, Plessa, E, Wlodarski, T, Ahn, M, Sidhu, H, Becker, T.A, Beckmann, R, Cabrita, L.D, Christodoulou, J. | | Deposit date: | 2022-04-25 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Modulating co-translational protein folding by rational design and ribosome engineering.
Nat Commun, 13, 2022
|
|
5XE9
 
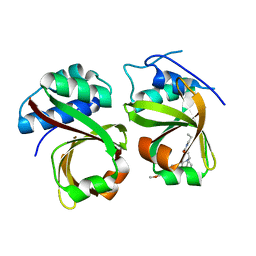 | | Crystal Structure of the Complex of the Peptidase Domain of Streptococcus mutans ComA with a Small Molecule Inhibitor. | | Descriptor: | Putative ABC transporter, ATP-binding protein ComA, [(1~{S},2~{R},4~{S},5~{R})-5-[5-(4-methoxyphenyl)-2-methyl-pyrazol-3-yl]-1-azabicyclo[2.2.2]octan-2-yl]methyl ~{N}-propylcarbamate | | Authors: | Ishii, S, Fukui, K, Yokoshima, S, Kumagai, K, Beniyama, Y, Kodama, T, Fukuyama, T, Okabe, T, Nagano, T, Kojima, H, Yano, T. | | Deposit date: | 2017-04-03 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | High-throughput Screening of Small Molecule Inhibitors of the Streptococcus Quorum-sensing Signal Pathway
Sci Rep, 7, 2017
|
|
7VI4
 
 | | Electron crystallographic structure of TIA-1 prion-like domain, A381T mutant | | Descriptor: | TIA-1 prion-like domain | | Authors: | Takaba, K, Maki-Yonekura, S, Sekiyama, N, Imamura, K, Kodama, T, Tochio, H, Yonekura, K. | | Deposit date: | 2021-09-24 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.95 Å) | | Cite: | ALS mutations in the TIA-1 prion-like domain trigger highly condensed pathogenic structures.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VI5
 
 | | Electron crystallographic structure of TIA-1 prion-like domain, wild type sequence | | Descriptor: | TIA-1 prion-like domain | | Authors: | Takaba, K, Maki-Yonekura, S, Sekiyama, N, Imamura, K, Kodama, T, Tochio, H, Yonekura, K. | | Deposit date: | 2021-09-24 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-29 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.761 Å) | | Cite: | ALS mutations in the TIA-1 prion-like domain trigger highly condensed pathogenic structures.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6IDP
 
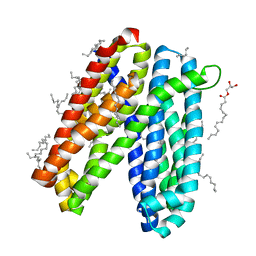 | | Crystal structure of Vibrio cholerae MATE transporter VcmN in the straight form | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, MATE family efflux transporter | | Authors: | Kusakizako, T, Claxton, D.P, Tanaka, Y, Maturana, A.D, Kuroda, T, Ishitani, R, Mchaourab, H.S, Nureki, O. | | Deposit date: | 2018-09-11 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | Structural Basis of H+-Dependent Conformational Change in a Bacterial MATE Transporter.
Structure, 27, 2019
|
|
5XE8
 
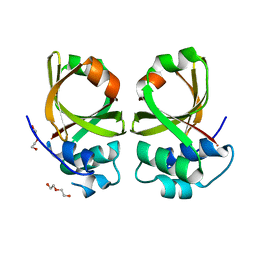 | | Crystal Structure of the Peptidase Domain of Streptococcus mutans ComA | | Descriptor: | DI(HYDROXYETHYL)ETHER, Putative ABC transporter, ATP-binding protein ComA | | Authors: | Ishii, S, Fukui, K, Yokoshima, S, Kumagai, K, Beniyama, Y, Kodama, T, Fukuyama, T, Okabe, T, Nagano, T, Kojima, H, Yano, T. | | Deposit date: | 2017-04-03 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | High-throughput Screening of Small Molecule Inhibitors of the Streptococcus Quorum-sensing Signal Pathway
Sci Rep, 7, 2017
|
|
7YPW
 
 | | Lloviu cuevavirus nucleoprotein-RNA complex | | Descriptor: | Nucleoprotein, RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3') | | Authors: | Hu, S.F, Fujita-Fujiharu, Y, Sugita, Y, Wendt, L, Muramoto, Y, Nakano, M, Hoenen, T, Noda, T. | | Deposit date: | 2022-08-04 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.0356 Å) | | Cite: | Cryoelectron microscopic structure of the nucleoprotein-RNA complex of the European filovirus, Lloviu virus.
Pnas Nexus, 2, 2023
|
|
7YR8
 
 | | Lloviu cuevavirus nucleoprotein(1-450 residues)-RNA complex | | Descriptor: | Nucleoprotein, RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3') | | Authors: | Hu, S.F, Fujita-Fujiharu, Y, Sugita, Y, Wendt, L, Muramoto, Y, Nakano, M, Hoenen, T, Noda, T. | | Deposit date: | 2022-08-09 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryoelectron microscopic structure of the nucleoprotein-RNA complex of the European filovirus, Lloviu virus.
Pnas Nexus, 2, 2023
|
|
6AVK
 
 | | Streptavidin bound to peptide-like compound KPM-6 | | Descriptor: | 1-hydroxydodecan-4-one, N-[(2H-1,3-benzodioxol-5-yl)methyl]-2-({[(2H-1,3-benzodioxol-5-yl)methyl][2-(chloromethyl)-1,3-oxazole-4-carbonyl]amino}methyl)-N-[(4-carbamoyl-1,3-oxazol-2-yl)methyl]-1,3-oxazole-4-carboxamide, Streptavidin | | Authors: | Park, H, Shamim, R, McEnaney, P, Kodadek, T. | | Deposit date: | 2017-09-02 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Efficient Workflow for Screening DNA-encoded One Bead One Compound Libraries Using a Flow Cytometer
To be Published
|
|
6IDS
 
 | | Crystal structure of Vibrio cholerae MATE transporter VcmN D35N mutant | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, MATE family efflux transporter | | Authors: | Kusakizako, T, Claxton, D.P, Tanaka, Y, Maturana, A.D, Kuroda, T, Ishitani, R, Mchaourab, H.S, Nureki, O. | | Deposit date: | 2018-09-11 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural Basis of H+-Dependent Conformational Change in a Bacterial MATE Transporter.
Structure, 27, 2019
|
|
6IDR
 
 | | Crystal structure of Vibrio cholerae MATE transporter VcmN in the bent form | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, MATE family efflux transporter | | Authors: | Kusakizako, T, Claxton, D.P, Tanaka, Y, Maturana, A.D, Kuroda, T, Ishitani, R, Mchaourab, H.S, Nureki, O. | | Deposit date: | 2018-09-11 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structural Basis of H+-Dependent Conformational Change in a Bacterial MATE Transporter.
Structure, 27, 2019
|
|
4ZHZ
 
 | | Endonuclease inhibitor bound to influenza strain H1N1 polymerase acidic subunit N-terminal region with expelling one of the metal ions in the active site | | Descriptor: | 5-(2-chlorobenzyl)-2-hydroxy-3-nitrobenzaldehyde, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Fudo, S, Yamamoto, N, Nukaga, M, Odagiri, T, Tashiro, M, Neya, S, Hoshino, T. | | Deposit date: | 2015-04-27 | | Release date: | 2015-05-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two Distinctive Binding Modes of Endonuclease Inhibitors to the N-Terminal Region of Influenza Virus Polymerase Acidic Subunit
Biochemistry, 55, 2016
|
|
4YYL
 
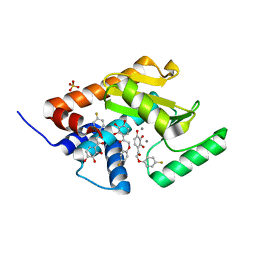 | | Phenolic acid derivative bound to influenza strain H1N1 polymerase subunit PA endonuclease | | Descriptor: | 2-(4-fluorophenoxy)-1-(2,3,4-trihydroxyphenyl)ethanone, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Fudo, S, Yamamoto, N, Nukaga, M, Odagiri, T, Tashiro, M, Neya, S, Hoshino, T. | | Deposit date: | 2015-03-24 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Structural and computational study on inhibitory compounds for endonuclease activity of influenza virus polymerase
Bioorg.Med.Chem., 23, 2015
|
|
7D7M
 
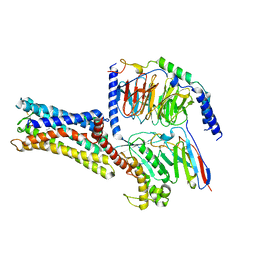 | | Cryo-EM Structure of the Prostaglandin E Receptor EP4 Coupled to G Protein | | Descriptor: | (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-cyclopentyl]hept-5-enoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Nojima, S, Fujita, Y, Kimura, T.K, Nomura, N, Suno, R, Morimoto, K, Yamamoto, M, Noda, T, Iwata, S, Shigematsu, H, Kobayashi, T. | | Deposit date: | 2020-10-05 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM Structure of the Prostaglandin E Receptor EP4 Coupled to G Protein.
Structure, 29, 2021
|
|
6K96
 
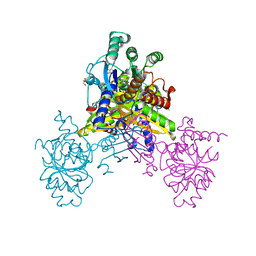 | | Crystal structure of Ari2 | | Descriptor: | Five-membered-cyclitol-phosphate synthase, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Miyanaga, A, Tsunoda, T, Kudo, F, Eguchi, T. | | Deposit date: | 2019-06-14 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Stereochemistry in the Reaction of themyo-Inositol Phosphate Synthase Ortholog Ari2 during Aristeromycin Biosynthesis.
Biochemistry, 58, 2019
|
|
4ZI0
 
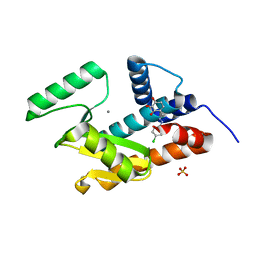 | | Endonuclease inhibitor bound to influenza strain H1N1 polymerase acidic subunit N-terminal region without a chelation to the metal ions in the active site | | Descriptor: | 4-{(E)-[2-(4-chlorophenyl)hydrazinylidene]methyl}benzene-1,2,3-triol, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Fudo, S, Yamamoto, N, Nukaga, M, Odagiri, T, Tashiro, M, Neya, S, Hoshino, T. | | Deposit date: | 2015-04-27 | | Release date: | 2015-05-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Two Distinctive Binding Modes of Endonuclease Inhibitors to the N-Terminal Region of Influenza Virus Polymerase Acidic Subunit
Biochemistry, 55, 2016
|
|
4ZQQ
 
 | | Apo form of influenza strain H1N1 polymerase acidic subunit N-terminal region | | Descriptor: | MANGANESE (II) ION, Polymerase acidic protein,Polymerase acidic protein, SULFATE ION | | Authors: | Fudo, S, Yamamoto, N, Nukaga, M, Odagiri, T, Tashiro, M, Neya, S, Hoshino, T. | | Deposit date: | 2015-05-11 | | Release date: | 2015-06-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Two Distinctive Binding Modes of Endonuclease Inhibitors to the N-Terminal Region of Influenza Virus Polymerase Acidic Subunit.
Biochemistry, 55, 2016
|
|
