1FZZ
 
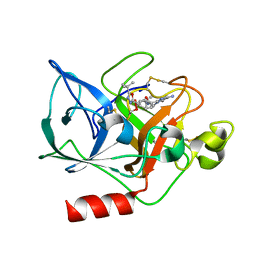 | | THE CRYSTAL STRUCTURE OF THE COMPLEX OF NON-PEPTIDIC INHIBITOR ONO-6818 AND PORCINE PANCREATIC ELASTASE. | | Descriptor: | 2-(5-AMINO-6-OXO-2-PHENYL-6H-PYRIMIDIN-1-YL)-N-[2-(5-TERT-BUTYL-1,3,4-OXADIAZOL-2-YL)-1-(METHYLETHYL)-2-HYDROXYETHYL]ACETAMIDE, ELASTASE 1 | | Authors: | Odagaki, Y, Ohmoto, K, Matsuoka, S, Hamanaka, N, Nakai, H, Toda, M, Katsuya, Y. | | Deposit date: | 2000-10-04 | | Release date: | 2001-10-04 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The crystal structure of the complex of non-peptidic inhibitor of human neutrophil elastase ONO-6818 and porcine pancreatic elastase.
Bioorg.Med.Chem., 9, 2001
|
|
4ZD6
 
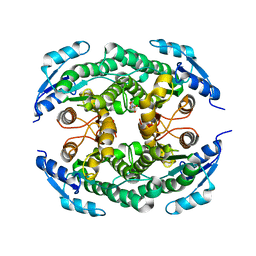 | | Halohydrin hydrogen-halide-lyase, HheB | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Halohydrin epoxidase B, ... | | Authors: | Watanabe, F, Yu, F, Ohtaki, A, Yamanaka, Y, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2015-04-17 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of halohydrin hydrogen-halide-lyases from Corynebacterium sp. N-1074
Proteins, 83, 2015
|
|
7XTX
 
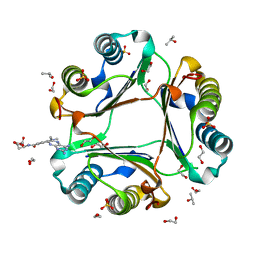 | | High resolution crystal structure of human macrophage migration inhibitory factor in complex with methotrexate | | Descriptor: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, Macrophage migration inhibitory factor, ... | | Authors: | Sugishima, K, Noguchi, K, Yohda, M, Odaka, M, Matsumura, H. | | Deposit date: | 2022-05-18 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Identification of methotrexate as an inhibitor of macrophage migration inhibitory factor by high-resolution crystal structure analysis
To Be Published
|
|
4Z9F
 
 | | Halohydrin hydrogen-halide-lyase, HheA | | Descriptor: | CHLORIDE ION, Halohydrin epoxidase A | | Authors: | Watanabe, F, Yu, F, Ohtaki, A, Yamanaka, Y, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2015-04-10 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of halohydrin hydrogen-halide-lyases from Corynebacterium sp. N-1074
Proteins, 83, 2015
|
|
5B19
 
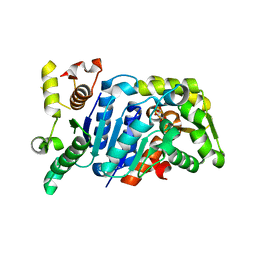 | | Picrophilus torridus aspartate racemase | | Descriptor: | Aspartate racemase, L(+)-TARTARIC ACID | | Authors: | Aihara, T, Ito, T, Yamanaka, Y, Noguchi, K, Odaka, M, Sekine, M, Homma, H, Yohda, M. | | Deposit date: | 2015-11-30 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Structural and functional characterization of aspartate racemase from the acidothermophilic archaeon Picrophilus torridus
Extremophiles, 20, 2016
|
|
5B4V
 
 | | Crystal structure of D-3-hydroxybutyrate dehydrogenase from Alcaligenes faecalis complexed with NAD+ and an inhibitor methylmalonate | | Descriptor: | 3-hydroxybutyrate dehydrogenase, CHLORIDE ION, METHYLMALONIC ACID, ... | | Authors: | Kanazawa, H, Tsunoda, M, Hoque, M.M, Suzuki, K, Yamamoto, T, Takenaka, A. | | Deposit date: | 2016-04-19 | | Release date: | 2016-08-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into the catalytic reaction trigger and inhibition of D-3-hydroxybutyrate dehydrogenase
Acta Crystallogr.,Sect.F, 72, 2016
|
|
7E1P
 
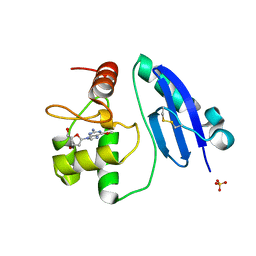 | | Crystal structure of Sulfurisphaera tokodaii O6-methylguanine methyltransferase C120S variant in complex with O6-methyldeoxyguanosine | | Descriptor: | (2~{R},3~{S},5~{R})-5-(2-azanyl-6-methoxy-purin-9-yl)-2-(hydroxymethyl)oxolan-3-ol, Methylated-DNA--protein-cysteine methyltransferase, SULFATE ION | | Authors: | Kikuchi, M, Yamauchi, T, Iizuka, Y, Tsunoda, M. | | Deposit date: | 2021-02-03 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Roles of the hydroxy group of tyrosine in crystal structures of Sulfurisphaera tokodaii O6-methylguanine-DNA methyltransferase.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7VMO
 
 | | Structure of recombinant RyR2 (Ca2+ dataset, class 1, open state) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ... | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations.
Nat Commun, 13, 2022
|
|
7VMP
 
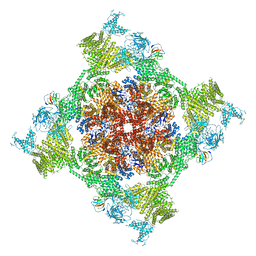 | | Structure of recombinant RyR2 (Ca2+ dataset, class 2, open state) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ... | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations.
Nat Commun, 13, 2022
|
|
7VMQ
 
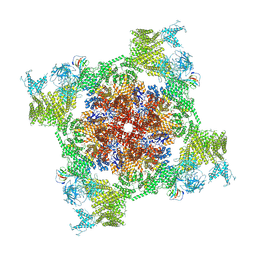 | | Structure of recombinant RyR2 (Ca2+ dataset, class 3, open state) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ... | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations
Nat Commun, 13, 2022
|
|
4DEN
 
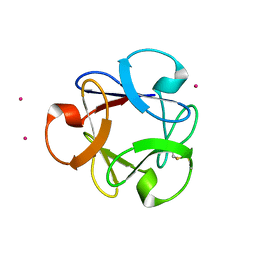 | | Structural insightsinto potent, specific anti-HIV property of actinohivin; Crystal structure of actinohivin in complex with alpha(1-2) mannobiose moiety of high-mannose type glycan of gp120 | | Descriptor: | Actinohivin, POTASSIUM ION, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose | | Authors: | Hoque, M.M, Suzuki, K, Tsunoda, M, Jiang, J, Zhang, F, Takahashi, A, Naomi, O, Zhang, X, Sekiguchi, T, Tanaka, H, Omura, S, Takenaka, A. | | Deposit date: | 2012-01-20 | | Release date: | 2012-11-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the specific anti-HIV property of actinohivin: structure of its complex with the alpha(1–2)mannobiose moiety of gp120
Acta Crystallogr.,Sect.D, 68, 2012
|
|
7VMS
 
 | | Structure of recombinant RyR2 mutant K4593A (Ca2+ dataset) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ... | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations
Nat Commun, 13, 2022
|
|
5B12
 
 | | Crystal structure of the B-type halohydrin hydrogen-halide-lyase mutant F71W/Q125T/D199H from Corynebacterium sp. N-1074 | | Descriptor: | CHLORIDE ION, Halohydrin epoxidase B | | Authors: | Watanabe, F, Yu, F, Ohtaki, A, Yamanaka, Y, Noguchi, K, Odaka, M, Yohda, M. | | Deposit date: | 2015-11-17 | | Release date: | 2016-08-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.721 Å) | | Cite: | Improvement of enantioselectivity of the B-type halohydrin hydrogen-halide-lyase from Corynebacterium sp. N-1074
J.Biosci.Bioeng., 122, 2016
|
|
4END
 
 | | Crystal structure of anti-HIV actinohivin in complex with alpha-1,2-mannobiose (P 2 21 21 form) | | Descriptor: | ACETONITRILE, Actinohivin, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose | | Authors: | Hoque, M.M, Suzuki, K, Tsunoda, M, Jiang, J, Zhang, F, Takahashi, A, Naomi, O, Zhang, X, Sekiguchi, T, Tanaka, H, Omura, S, Takenaka, A. | | Deposit date: | 2012-04-13 | | Release date: | 2013-07-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Matured structure of anti-HIV lectin actinohivin in complex with 1,2-mannobiose
To be Published
|
|
4P6A
 
 | | Crystal structure of a potent anti-HIV lectin actinohivin in complex with alpha-1,2-mannotriose | | Descriptor: | Actinohivin, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose | | Authors: | Zhang, F, Hoque, M.M, Suzuki, K, Tsunoda, M, Naomi, O, Tanaka, H, Takenaka, A. | | Deposit date: | 2014-03-23 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | The characteristic structure of anti-HIV actinohivin in complex with three HMTG D1 chains of HIV-gp120.
Chembiochem, 15, 2014
|
|
2AHJ
 
 | | NITRILE HYDRATASE COMPLEXED WITH NITRIC OXIDE | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, FE (III) ION, NITRIC OXIDE, ... | | Authors: | Nagashima, S, Nakasako, M, Dohmae, N, Tsujimura, M, Takio, K, Odaka, M, Yohda, M, Kamiya, N, Endo, I. | | Deposit date: | 1997-12-24 | | Release date: | 1999-01-27 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Novel non-heme iron center of nitrile hydratase with a claw setting of oxygen atoms.
Nat.Struct.Biol., 5, 1998
|
|
3VDR
 
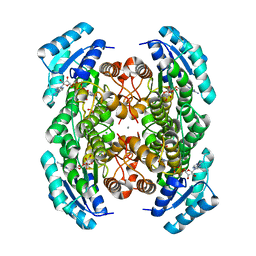 | | Crystal structure of D-3-hydroxybutyrate dehydrogenase, prepared in the presence of the substrate D-3-hydroxybutyrate and NAD(+) | | Descriptor: | (3R)-3-hydroxybutanoic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ACETOACETIC ACID, ... | | Authors: | Hoque, M.M, Shimizu, S, Juan, E.C.M, Sato, Y, Hossain, M.T, Yamamoto, T, Imamura, S, Amano, H, Suzuki, K, Sekiguchi, T, Tsunoda, M, Takenaka, A. | | Deposit date: | 2012-01-06 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of D-3-hydroxybutyrate dehydrogenase prepared in the presence of the substrate D-3-hydroxybutyrate and NAD+.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
5AXE
 
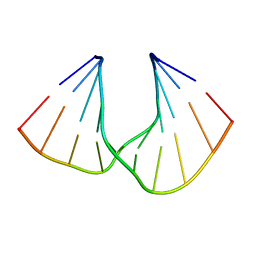 | | Crystal Structure Analysis of DNA Duplexes containing sulfoamide-bridged nucleic acid (SuNA-NH) | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(LSH)P*AP*CP*GP*C)-3') | | Authors: | Mitsuoka, Y, Aoyama, H, Kugimiya, A, Fujimura, Y, Yamamoto, T, Waki, R, Wada, F, Tahara, S, Sawamura, M, Noda, M, Hari, Y, Obika, S. | | Deposit date: | 2015-07-28 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Effect of an N-substituent in sulfonamide-bridged nucleic acid (SuNA) on hybridization ability and duplex structure.
Org.Biomol.Chem., 14, 2016
|
|
5AXF
 
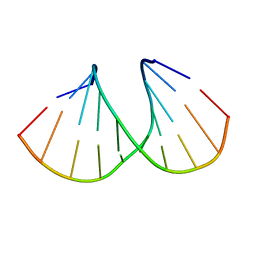 | | Crystal Structure Analysis of DNA Duplexes containing sulfoamide-bridged nucleic acid (SuNA-NMe) | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*AP*(LSM)P*AP*CP*GP*C)-3') | | Authors: | Mitsuoka, Y, Aoyama, H, Kugimiya, A, Fujimura, Y, Yamamoto, T, Waki, R, Wada, F, Tahara, S, Sawamura, M, Noda, M, Hari, Y, Obika, S. | | Deposit date: | 2015-07-28 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Effect of an N-substituent in sulfonamide-bridged nucleic acid (SuNA) on hybridization ability and duplex structure.
Org.Biomol.Chem., 14, 2016
|
|
3IS1
 
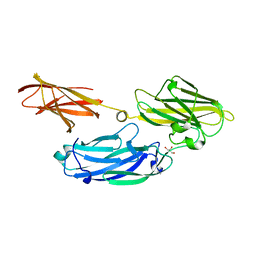 | | Crystal structure of functional region of UafA from Staphylococcus saprophyticus in C2 form at 2.45 angstrom resolution | | Descriptor: | GLYCEROL, Uro-adherence factor A | | Authors: | Tanaka, Y, Matsuoka, E, Shouji, Y, Kuroda, M, Tanaka, I, Yao, M. | | Deposit date: | 2009-08-24 | | Release date: | 2010-09-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of the functional region of Uro-adherence factor A from Staphylococcus saprophyticus reveals participation of the B domain in ligand binding
Protein Sci., 20, 2011
|
|
1WTG
 
 | | Human Factor Viia-Tissue Factor Complexed with ethylsulfonamide-D-biphenylalanine-Gln-p-aminobenzamidine | | Descriptor: | 2-(3-BIPHENYL-4-YL-2-ETHANESULFONYLAMINO-PROPIONYLAMINO)-PENTANEDIOIC ACID 5-AMIDE 1-(4-CARBAMIMIDOYL-BENZYLAMIDE), CALCIUM ION, Coagulation factor VII, ... | | Authors: | Kadono, S, Sakamoto, S, Kikuchi, Y, Oh-Eda, M, Yabuta, N, Kitazawa, K, Yoshihashi, T, Suzuki, T, Koga, T, Hattori, K, Shiraishi, T, Kodama, M, Haramura, H, Ono, Y, Esaki, T, Sato, H, Watanabe, Y, Itoh, S, Ohta, M, Kozono, T. | | Deposit date: | 2004-11-23 | | Release date: | 2005-11-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel interactions of large P3 moiety and small P4 moiety in the binding of the peptide mimetic factor VIIa inhibitor
Biochem.Biophys.Res.Commun., 326, 2005
|
|
3IS0
 
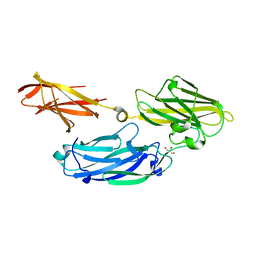 | | Crystal structure of functional region of UafA from Staphylococcus saprophyticus in the presence of cholesterol | | Descriptor: | GLYCEROL, Uro-adherence factor A | | Authors: | Tanaka, Y, Shouji, Y, Matsuoka, E, Kuroda, M, Tanaka, I, Yao, M. | | Deposit date: | 2009-08-24 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Functional Region of UafA from Staphylococcus saprophyticus
To be Published
|
|
3IRP
 
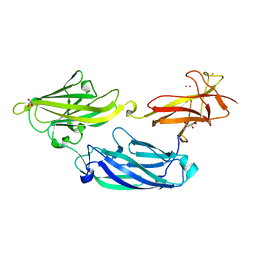 | | Crystal structure of functional region of UafA from Staphylococcus saprophyticus at 1.50 angstrom resolution | | Descriptor: | GLYCEROL, POTASSIUM ION, Uro-adherence factor A | | Authors: | Tanaka, Y, Shouji, Y, Matsuoka, E, Kuroda, M, Tanaka, I, Yao, M. | | Deposit date: | 2009-08-24 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the functional region of Uro-adherence factor A from Staphylococcus saprophyticus reveals participation of the B domain in ligand binding
Protein Sci., 20, 2011
|
|
3IRZ
 
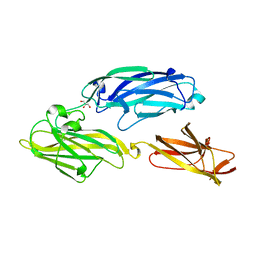 | | Crystal structure of functional region of UafA from Staphylococcus saprophyticus in P212121 form | | Descriptor: | GLYCEROL, Uro-adherence factor A | | Authors: | Tanaka, Y, Shouji, Y, Matsuoka, E, Kuroda, M, Tanaka, I, Yao, M. | | Deposit date: | 2009-08-24 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the functional region of Uro-adherence factor A from Staphylococcus saprophyticus reveals participation of the B domain in ligand binding
Protein Sci., 20, 2011
|
|
6J28
 
 | | Crystal structure of the branched-chain polyamine synthase C9 mutein from Thermus thermophilus (Tth-BpsA C9) in complex with N4-aminopropylspermidine and 5'-methylthioadenosine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-DEOXY-5'-METHYLTHIOADENOSINE, FE (III) ION, ... | | Authors: | Mizohata, E, Toyoda, M, Fujita, J, Inoue, T. | | Deposit date: | 2018-12-31 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The C-terminal flexible region of branched-chain polyamine synthase facilitates substrate specificity and catalysis.
Febs J., 286, 2019
|
|
