3JUB
 
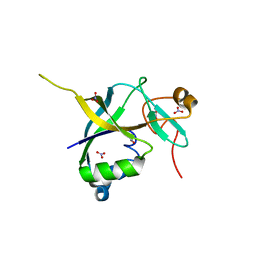 | | Human gamma-glutamylamine cyclotransferase | | Descriptor: | AIG2-like domain-containing protein 1, NITRATE ION | | Authors: | Oakley, A.J. | | Deposit date: | 2009-09-15 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Identification and characterization of {gamma}-glutamylamine cyclotransferase: An enzyme responsible for {gamma}-glutamyl-{epsilon}-lysine catabolism
J.Biol.Chem., 285, 2010
|
|
6CBS
 
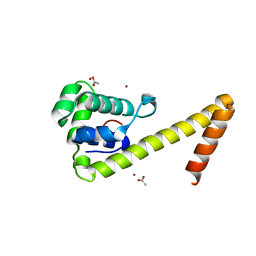 | |
6CBR
 
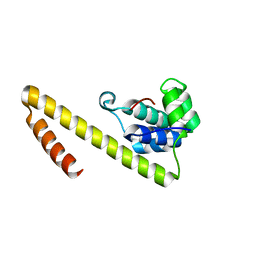 | |
1IGO
 
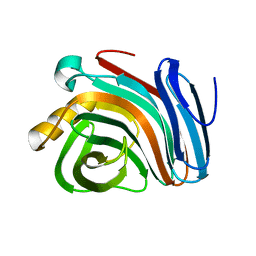 | | Family 11 xylanase | | Descriptor: | SULFATE ION, family 11 xylanase | | Authors: | Oakley, A.J, Thomson, C, Heinrich, T, Dunlop, R, Wilce, M.C.J. | | Deposit date: | 2001-04-18 | | Release date: | 2002-04-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Characterization of a family 11 xylanase from Bacillus subtillis B230 used for paper bleaching.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
18GS
 
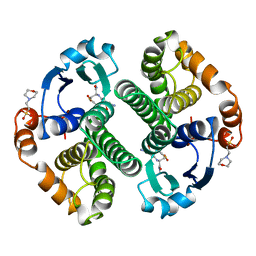 | | GLUTATHIONE S-TRANSFERASE P1-1 COMPLEXED WITH 1-(S-GLUTATHIONYL)-2,4-DINITROBENZENE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE S-(2,4 DINITROBENZENE), GLUTATHIONE S-TRANSFERASE | | Authors: | Oakley, A.J, Lo Bello, M, Ricci, G, Federici, G, Parker, M.W. | | Deposit date: | 1997-12-07 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The ligandin (non-substrate) binding site of human Pi class glutathione transferase is located in the electrophile binding site (H-site).
J.Mol.Biol., 291, 1999
|
|
11GS
 
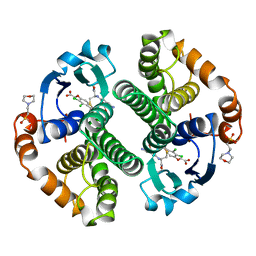 | | Glutathione s-transferase complexed with ethacrynic acid-glutathione conjugate (form ii) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ETHACRYNIC ACID, GLUTATHIONE, ... | | Authors: | Oakley, A.J, Lo Bello, M, Mazzetti, A.P, Federici, G, Parker, M.W. | | Deposit date: | 1997-11-03 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The glutathione conjugate of ethacrynic acid can bind to human pi class glutathione transferase P1-1 in two different modes.
FEBS Lett., 419, 1997
|
|
16GS
 
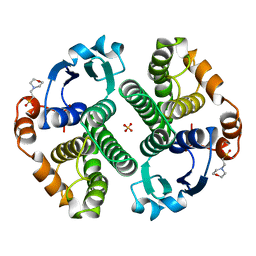 | | GLUTATHIONE S-TRANSFERASE P1-1 APO FORM 3 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE S-TRANSFERASE, SULFATE ION | | Authors: | Oakley, A.J, Lo Bello, M, Ricci, G, Federici, G, Parker, M.W. | | Deposit date: | 1997-11-30 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evidence for an induced-fit mechanism operating in pi class glutathione transferases.
Biochemistry, 37, 1998
|
|
14GS
 
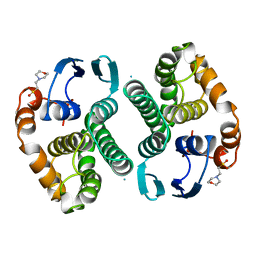 | | GLUTATHIONE S-TRANSFERASE P1-1 APO FORM 1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE S-TRANSFERASE | | Authors: | Oakley, A.J, Lo Bello, M, Ricci, G, Federici, G, Parker, M.W. | | Deposit date: | 1997-11-29 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Evidence for an induced-fit mechanism operating in pi class glutathione transferases.
Biochemistry, 37, 1998
|
|
17GS
 
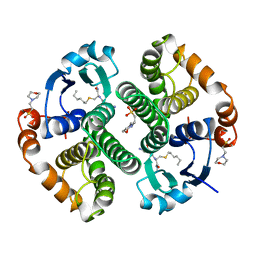 | | GLUTATHIONE S-TRANSFERASE P1-1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE S-TRANSFERASE, S-HEXYLGLUTATHIONE | | Authors: | Oakley, A.J, Lo Bello, M, Parker, M.W. | | Deposit date: | 1997-12-07 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Glutathione S-transferase P1-1
To be published
|
|
3IBT
 
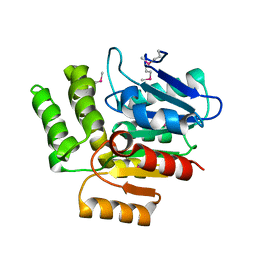 | | Structure of 1H-3-hydroxy-4-oxoquinoline 2,4-dioxygenase (QDO) | | Descriptor: | 1H-3-hydroxy-4-oxoquinoline 2,4-dioxygenase | | Authors: | Oakley, A.J. | | Deposit date: | 2009-07-16 | | Release date: | 2010-02-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for cofactor-independent dioxygenation of N-heteroaromatic compounds at the alpha/beta-hydrolase fold.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3K4Q
 
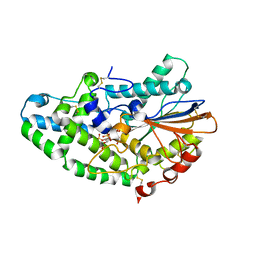 | | Aspergillus niger Phytase in complex with myo-inositol hexakis sulfate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-phytase A, D-MYO-INOSITOL-HEXASULPHATE | | Authors: | Oakley, A.J. | | Deposit date: | 2009-10-06 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of Aspergillus niger phytase PhyA in complex with a phytate mimetic
Biochem.Biophys.Res.Commun., 397, 2010
|
|
3K37
 
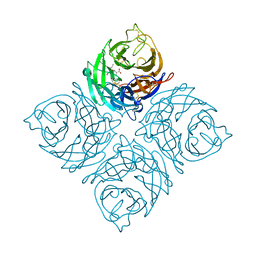 | | Crystal Structure of B/Perth Neuraminidase in complex with Peramivir | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(1-ACETYLAMINO-2-ETHYL-BUTYL)-4-GUANIDINO-2-HYDROXY-CYCLOPENTANECARBOXYLIC ACID, CALCIUM ION, ... | | Authors: | Oakley, A.J, McKimm-Breschkin, J.L. | | Deposit date: | 2009-10-02 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Basis of Resistance to Neuraminidase Inhibitors of Influenza B Viruses.
J.Med.Chem., 2010
|
|
3K39
 
 | |
3K38
 
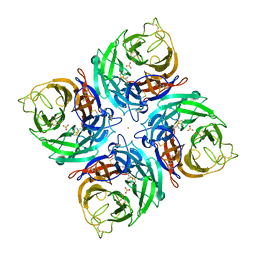 | |
3K3A
 
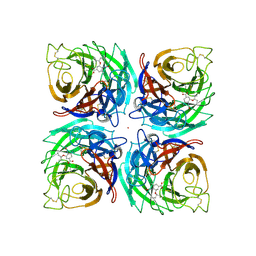 | |
3K36
 
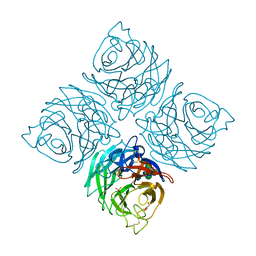 | |
3K4P
 
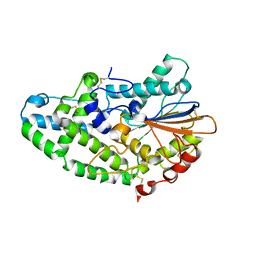 | | Aspergillus niger Phytase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-phytase A | | Authors: | Oakley, A.J. | | Deposit date: | 2009-10-06 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of Aspergillus niger phytase PhyA in complex with a phytate mimetic
Biochem.Biophys.Res.Commun., 397, 2010
|
|
4MZ9
 
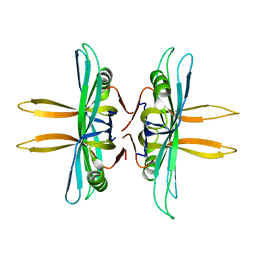 | | Revised structure of E. coli SSB | | Descriptor: | Single-stranded DNA-binding protein | | Authors: | Oakley, A.J. | | Deposit date: | 2013-09-29 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Intramolecular binding mode of the C-terminus of Escherichia coli single-stranded DNA binding protein determined by nuclear magnetic resonance spectroscopy.
Nucleic Acids Res., 42, 2014
|
|
3VLN
 
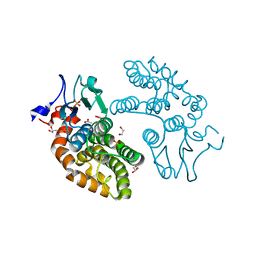 | | Human Glutathione Transferase O1-1 C32S Mutant in Complex with Ascorbic Acid | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ASCORBIC ACID, ... | | Authors: | Brock, J, Board, P.G, Oakley, A.J. | | Deposit date: | 2011-12-02 | | Release date: | 2012-05-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the dehydroascorbate reductase activity of human omega-class glutathione transferases.
J.Mol.Biol., 420, 2012
|
|
4ZW2
 
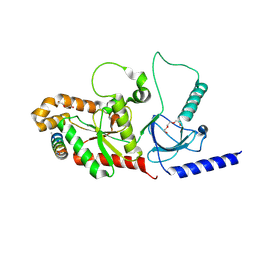 | |
4GX9
 
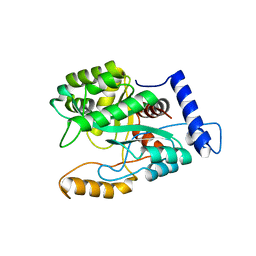 | | Crystal structure of a DNA polymerase III alpha-epsilon chimera | | Descriptor: | DNA polymerase III subunit epsilon,DNA polymerase III subunit alpha | | Authors: | Li, N, Horan, N, Xu, Z.-Q, Jacques, D, Dixon, N.E, Oakley, A.J. | | Deposit date: | 2012-09-04 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Proofreading exonuclease on a tether: the complex between the E. coli DNA polymerase III subunits alpha, {varepsilon}, theta and beta reveals a highly flexible arrangement of the proofreading domain
Nucleic Acids Res., 41, 2013
|
|
1EOG
 
 | | CRYSTAL STRUCTURE OF PI CLASS GLUTATHIONE TRANSFERASE | | Descriptor: | GLUTATHIONE S-TRANSFERASE | | Authors: | Rossjohn, J, McKinstry, W.J, Oakley, A.J, Parker, M.W, Stenberg, G, Mannervik, B, Dragani, B, Cocco, R, Aceto, A. | | Deposit date: | 2000-03-22 | | Release date: | 2000-10-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of thermolabile mutants of human glutathione transferase P1-1.
J.Mol.Biol., 302, 2000
|
|
1EOH
 
 | | GLUTATHIONE TRANSFERASE P1-1 | | Descriptor: | GLUTATHIONE S-TRANSFERASE | | Authors: | Rossjohn, J, McKinstry, W.J, Oakley, A.J, Parker, M.W, Stenberg, G, Mannervik, B, Dragani, B, Cocco, R, Aceto, A. | | Deposit date: | 2000-03-22 | | Release date: | 2000-10-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of thermolabile mutants of human glutathione transferase P1-1.
J.Mol.Biol., 302, 2000
|
|
4UY3
 
 | | Cytoplasmic domain of bacterial cell division protein ezra | | Descriptor: | SEPTATION RING FORMATION REGULATOR EZRA | | Authors: | Cleverley, R.M, Barrett, J.R, Basle, A, Khai-Bui, N, Hewitt, L, Solovyova, A, Xu, Z, Daniela, R.A, Dixon, N.E, Harry, E.J, Oakley, A.J, Vollmer, W, Lewis, R.J. | | Deposit date: | 2014-08-28 | | Release date: | 2014-10-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Function of a Spectrin-Like Regulator of Bacterial Cytokinesis.
Nat.Commun., 5, 2014
|
|
4UXV
 
 | | Cytoplasmic domain of bacterial cell division protein EzrA | | Descriptor: | SEPTATION RING FORMATION REGULATOR EZRA | | Authors: | Cleverley, R.M, Barrett, J.R, Basle, A, Khai-Bui, N, Hewitt, L, Solovyova, A, Xu, Z, Daniela, R.A, Dixon, N.E, Harry, E.J, Oakley, A.J, Vollmer, W, Lewis, R.J. | | Deposit date: | 2014-08-27 | | Release date: | 2014-10-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.961 Å) | | Cite: | Structure and Function of a Spectrin-Like Regulator of Bacterial Cytokinesis.
Nat.Commun., 5, 2014
|
|
