1SL5
 
 | | Crystal Structure of DC-SIGN carbohydrate recognition domain complexed with LNFP III (Dextra L504). | | Descriptor: | CALCIUM ION, MAGNESIUM ION, alpha-L-fucopyranose-(1-3)-[beta-D-galactopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose, ... | | Authors: | Guo, Y, Feinberg, H, Conroy, E, Mitchell, D.A, Alvarez, R, Blixt, O, Taylor, M.E, Weis, W.I, Drickamer, K. | | Deposit date: | 2004-03-05 | | Release date: | 2004-06-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for distinct ligand-binding and targeting properties of the receptors
DC-SIGN and DC-SIGNR
Nat.Struct.Mol.Biol., 11, 2004
|
|
1RXX
 
 | | Structure of arginine deiminase | | Descriptor: | Arginine deiminase | | Authors: | Galkin, A, Kulakova, L, Sarikaya, E, Lim, K, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2003-12-18 | | Release date: | 2004-01-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural insight into arginine degradation by arginine deiminase, an antibacterial and parasite drug target.
J.Biol.Chem., 279, 2004
|
|
1SL4
 
 | | Crystal Structure of DC-SIGN carbohydrate recognition domain complexed with Man4 | | Descriptor: | CALCIUM ION, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose, mDC-SIGN1B type I isoform | | Authors: | Guo, Y, Feinberg, H, Conroy, E, Mitchell, D.A, Alvarez, R, Blixt, O, Taylor, M.E, Weis, W.I, Drickamer, K. | | Deposit date: | 2004-03-05 | | Release date: | 2004-06-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for distinct ligand-binding and targeting properties of the receptors
DC-SIGN and DC-SIGNR
Nat.Struct.Mol.Biol., 11, 2004
|
|
1SM7
 
 | | Solution structure of the recombinant pronapin precursor, BnIb. | | Descriptor: | recombinant Ib pronapin | | Authors: | Pantoja-Uceda, D, Palomares, O, Bruix, M, Villalba, M, Rodriguez, R, Rico, M, Santoro, J. | | Deposit date: | 2004-03-08 | | Release date: | 2005-02-01 | | Last modified: | 2013-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and stability against digestion of rproBnIb, a recombinant 2S albumin from rapeseed: relationship to its allergenic properties.
Biochemistry, 43, 2004
|
|
3PCQ
 
 | | Femtosecond X-ray protein Nanocrystallography | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Chapman, H.N, Fromme, P, Barty, A, White, T.A, Kirian, R.A, Aquila, A, Hunter, M.S, Schulz, J, Deponte, D.P, Weierstall, U, Doak, R.B, Maia, F.R.N.C, Martin, A.V, Schlichting, I, Lomb, L, Coppola, N, Shoeman, R.L, Epp, S.W, Hartmann, R, Rolles, D, Rudenko, A, Foucar, L, Kimmel, N, Weidenspointner, G, Holl, P, Liang, M, Barthelmess, M, Caleman, C, Boutet, S, Bogan, M.J, Krzywinski, J, Bostedt, C, Bajt, S, Gumprecht, L, Rudek, B, Erk, B, Schmidt, C, Homke, A, Reich, C, Pietschner, D, Struder, L, Hauser, G, Gorke, H, Ullrich, J, Herrmann, S, Schaller, G, Schopper, F, Soltau, H, Kuhnel, K.-U, Messerschmidt, M, Bozek, J.D, Hau-Riege, S.P, Frank, M, Hampton, C.Y, Sierra, R, Starodub, D, Williams, G.J, Hajdu, J, Timneanu, N, Seibert, M.M, Andreasson, J, Rocker, A, Jonsson, O, Svenda, M, Stern, S, Nass, K, Andritschke, R, Schroter, C.-D, Krasniqi, F, Bott, M, Schmidt, K.E, Wang, X, Grotjohann, I, Holton, J.M, Barends, T.R.M, Neutze, R, Marchesini, S, Fromme, R, Schorb, S, Rupp, D, Adolph, M, Gorkhover, T, Andersson, I, Hirsemann, H, Potdevin, G, Graafsma, H, Nilsson, B, Spence, J.C.H. | | Deposit date: | 2010-10-21 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (8.984 Å) | | Cite: | Femtosecond X-ray protein nanocrystallography.
Nature, 470, 2011
|
|
1PWL
 
 | | Crystal structure of human Aldose Reductase complexed with NADP and Minalrestat | | Descriptor: | 2[4-BROMO-2-FLUOROPHENYL)METHYL]-6-FLUOROSPIRO[ISOQUINOLINE-4-(1H),3'-PYRROLIDINE]-1,2',3,5'(2H)-TETRONE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, aldose reductase | | Authors: | El-Kabbani, O, Darmanin, C, Schneider, T.R, Hazemann, I, Ruiz, F, Oka, M, Joachimiak, A, Schulze-Briese, C, Tomizaki, T, Mitschler, A, Podjarny, A. | | Deposit date: | 2003-07-02 | | Release date: | 2004-02-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Ultrahigh resolution drug design. II. Atomic resolution structures of human aldose reductase holoenzyme complexed with Fidarestat and Minalrestat: implications for the binding of cyclic imide inhibitors
PROTEINS, 55, 2004
|
|
1PVT
 
 | | Crystal structure of sugar-phosphate aldolase from Thermotoga maritima | | Descriptor: | sugar-phosphate aldolase | | Authors: | Osipiuk, J, Cuff, M.E, Korolev, O, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-06-28 | | Release date: | 2003-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of sugar-phosphate aldolase from Thermotoga maritima.
To be Published
|
|
1TE4
 
 | | Solution structure of MTH187. Ontario Centre for Structural Proteomics target MTH0187_1_111; Northeast Structural Genomics Target TT740 | | Descriptor: | conserved protein MTH187 | | Authors: | Gignac, I, Julien, O, Yee, A, Arrowsmith, C.H, Gagne, S.M, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-05-24 | | Release date: | 2004-07-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | MTH187 from Methanobacterium thermoautotrophicum has three HEAT-like Repeats.
J.Biomol.Nmr, 35, 2006
|
|
1SUF
 
 | | Carbon Monoxide Dehydrogenase from Carboxydothermus hydrogenoformans-Inactive state | | Descriptor: | Carbon Monoxide Dehydrogenase 2, FE(4)-NI(1)-S(5) CLUSTER, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Dobbek, H, Svetlitchnyi, V, Liss, J, Meyer, O. | | Deposit date: | 2004-03-26 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Carbon Monoxide Induced Decomposition of the Active Site [Ni-4Fe-5S] Cluster of CO Dehydrogenase
J.Am.Chem.Soc., 126, 2004
|
|
1T2O
 
 | | Crystal structure of Se-SrtA, C184-Ala | | Descriptor: | sortase | | Authors: | Zong, Y, Bice, T.W, Ton-That, H, Schneewind, O, Narayana, S.V. | | Deposit date: | 2004-04-22 | | Release date: | 2004-09-07 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of Staphylococcus aureus sortase A and its substrate complex
J.Biol.Chem., 279, 2004
|
|
1T84
 
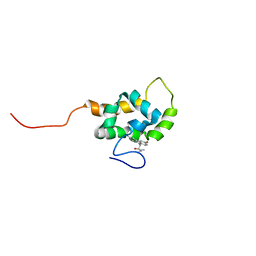 | | Solution structure of the Wiskott-Aldrich Syndrome Protein (WASP) autoinhibited core domain complexed with (S)-wiskostatin, a small molecule inhibitor | | Descriptor: | (2S)-1-(3,6-DIBROMO-9H-CARBAZOL-9-YL)-3-(DIMETHYLAMINO)PROPAN-2-OL, Wiskott-Aldrich syndrome protein | | Authors: | Peterson, J.R, Bickford, L.C, Morgan, D, Kim, A.S, Ouerfelli, O, Kirschner, M.W, Rosen, M.K. | | Deposit date: | 2004-05-11 | | Release date: | 2004-07-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Chemical inhibition of N-WASP by stabilization of a native autoinhibited conformation.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1TEG
 
 | | Crystal structure of the spinach plastocyanin mutants G8D/K30C/T69C and K30C/T69C- a study of the effect on crystal packing and thermostability from the introduction of a novel disulfide bond | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Plastocyanin, ... | | Authors: | Okvist, M, Jacobson, F, Jansson, H, Hansson, O, Sjolin, L. | | Deposit date: | 2004-05-25 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Novel Disulfide Bonds Effect the Thermostability of Plastocyanin. Crystal structures of the triple plastocyanin
mutant G8D/K30C/T69C and the double plastocyanin mutant K30C/T69C from spinach at 1.90 A and 1.96 A resolution, respectively.
To be Published
|
|
1SSN
 
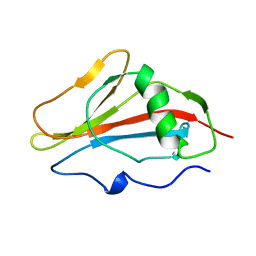 | | STAPHYLOKINASE, SAKSTAR VARIANT, NMR, 20 STRUCTURES | | Descriptor: | STAPHYLOKINASE | | Authors: | Ohlenschlager, O, Ramachandran, R, Guhrs, K.H, Schlott, B, Brown, L.R. | | Deposit date: | 1998-06-07 | | Release date: | 1998-12-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance solution structure of the plasminogen-activator protein staphylokinase.
Biochemistry, 37, 1998
|
|
1SU8
 
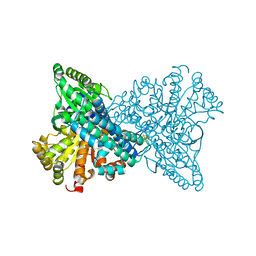 | | Carbon Monoxide Induced Decomposition of the Active Site [Ni-4Fe-5S] Cluster of CO Dehydrogenase | | Descriptor: | Carbon monoxide dehydrogenase 2, FE(4)-NI(1)-S(5) CLUSTER, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Dobbek, H, Svetlitchnyi, V, Liss, J, Meyer, O. | | Deposit date: | 2004-03-26 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Carbon Monoxide Induced Decomposition of the Active Site [Ni-4Fe-5S] Cluster of CO Dehydrogenase
J.Am.Chem.Soc., 126, 2004
|
|
1T2W
 
 | | Crystal Structure of Sortase A in Complex with a LPETG peptide | | Descriptor: | Class A sortase SrtA, Peptide LEU-PRO-GLU-THR-GLY | | Authors: | Zong, Y, Bice, T.W, Ton-That, H, Schneewind, O, Narayana, S.V. | | Deposit date: | 2004-04-23 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Staphylococcus aureus sortase A and its substrate complex
J.Biol.Chem., 279, 2004
|
|
1QJV
 
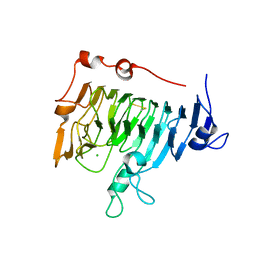 | | Pectin methylesterase PemA from Erwinia chrysanthemi | | Descriptor: | CHLORIDE ION, PECTIN METHYLESTERASE | | Authors: | Jenkins, J, Mayans, O, Smith, D, Worboys, K, Pickersgill, R. | | Deposit date: | 1999-07-05 | | Release date: | 2000-07-14 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Three-Dimensional Structure of Erwinia Chrysanthemi Pectin Methylesterase Reveals a Novel Esterase Active Site
J.Mol.Biol., 305, 2001
|
|
1QQT
 
 | | METHIONYL-TRNA SYNTHETASE FROM ESCHERICHIA COLI | | Descriptor: | METHIONYL-TRNA SYNTHETASE, ZINC ION | | Authors: | Mechulam, Y, Schmitt, E, Maveyraud, L, Zelwer, C, Nureki, O, Yokoyama, S, Konno, M, Blanquet, S. | | Deposit date: | 1999-06-08 | | Release date: | 2000-01-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of Escherichia coli methionyl-tRNA synthetase highlights species-specific features.
J.Mol.Biol., 294, 1999
|
|
2VCB
 
 | | Family 89 Glycoside Hydrolase from Clostridium perfringens in complex with PUGNAc | | Descriptor: | ALPHA-N-ACETYLGLUCOSAMINIDASE, CALCIUM ION, O-(2-ACETAMIDO-2-DEOXY D-GLUCOPYRANOSYLIDENE) AMINO-N-PHENYLCARBAMATE | | Authors: | Ficko-Blean, E, Stubbs, K.A, Berg, O, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2007-09-19 | | Release date: | 2008-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Mechanistic Insight Into the Basis of Mucopolysaccharidosis Iiib.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2VCC
 
 | | Family 89 Glycoside Hydrolase from Clostridium perfringens | | Descriptor: | ALPHA-N-ACETYLGLUCOSAMINIDASE, CALCIUM ION, GLYCEROL, ... | | Authors: | Ficko-Blean, E, Stubbs, K.A, Berg, O, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2007-09-19 | | Release date: | 2008-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Mechanistic Insight Into the Basis of Mucopolysaccharidosis Iiib.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
1RFR
 
 | | NMR structure of the 30mer stemloop-D of coxsackieviral RNA | | Descriptor: | stemloop-D RNA of the 5'-cloverleaf of coxsackievirus B3 | | Authors: | Ohlenschlager, O, Wohnert, J, Bucci, E, Seitz, S, Hafner, S, Ramachandran, R, Zell, R, Gorlach, M. | | Deposit date: | 2003-11-10 | | Release date: | 2004-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of the stemloop D subdomain of coxsackievirus B3 cloverleaf
RNA and its interaction with the proteinase 3C.
STRUCTURE, 12, 2004
|
|
1RLW
 
 | |
1SUA
 
 | | SUBTILISIN BPN' | | Descriptor: | SUBTILISIN BPN', TETRAPEPTIDE ALA-LEU-ALA-LEU | | Authors: | Almog, O, Gilliland, G.L. | | Deposit date: | 1997-01-14 | | Release date: | 1998-01-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of calcium-independent subtilisin BPN' with restored thermal stability folded without the prodomain.
Proteins, 31, 1998
|
|
1TFF
 
 | | Structure of Otubain-2 | | Descriptor: | Ubiquitin thiolesterase protein OTUB2 | | Authors: | Nanao, M.H, Tcherniuk, S.O, Chroboczek, J, Dideberg, O, Dessen, A, Balakirev, M.Y. | | Deposit date: | 2004-05-27 | | Release date: | 2004-08-03 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human otubain 2.
Embo Rep., 5, 2004
|
|
1SU7
 
 | | Carbon Monoxide Dehydrogenase from Carboxydothermus hydrogenoformans- DTT reduced state | | Descriptor: | Carbon monoxide dehydrogenase 2, FE(4)-NI(1)-S(5) CLUSTER, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Dobbek, H, Svetlitchnyi, V, Liss, J, Meyer, O. | | Deposit date: | 2004-03-26 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Carbon Monoxide Induced Decomposition of the Active Site [Ni-4Fe-5S] Cluster of CO Dehydrogenase
J.Am.Chem.Soc., 126, 2004
|
|
2RJM
 
 | | 3Ig structure of titin domains I67-I69 E-to-A mutated variant | | Descriptor: | Titin | | Authors: | von Castelmur, E, Marino, M, Labeit, D, Labeit, S, Mayans, O. | | Deposit date: | 2007-10-15 | | Release date: | 2008-01-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A regular pattern of Ig super-motifs defines segmental flexibility as the elastic mechanism of the titin chain
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
