3NGH
 
 | | Molecular Analysis of the Interaction of the HDL Receptor SR-BI with the Adaptor Protein PDZK1 | | Descriptor: | PDZ domain-containing protein 1 | | Authors: | Kocher, O, Birrane, G, Krieger, M, Ladias, J.A. | | Deposit date: | 2010-06-11 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In vitro and in vivo analysis of the binding of the C terminus of the HDL receptor scavenger receptor class B, type I (SR-BI), to the PDZ1 domain of its adaptor protein PDZK1.
J.Biol.Chem., 285, 2010
|
|
7R4W
 
 | | Single stranded DNA binding protein SSB M5 from Fervidobacterium gondwanense | | Descriptor: | ACETIC ACID, PHOSPHATE ION, Single-stranded DNA-binding protein | | Authors: | Hakansson, M, Svensson, L.A, Werbowy, O, Al-Karadaghi, S, Kaczorowski, T, Kaczorowska, A.K, Dorawa, S. | | Deposit date: | 2022-02-09 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular characterization of a single stranded DNA binding protein from Fervidobacterium gondwanense
To Be Published
|
|
6IDP
 
 | | Crystal structure of Vibrio cholerae MATE transporter VcmN in the straight form | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, MATE family efflux transporter | | Authors: | Kusakizako, T, Claxton, D.P, Tanaka, Y, Maturana, A.D, Kuroda, T, Ishitani, R, Mchaourab, H.S, Nureki, O. | | Deposit date: | 2018-09-11 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | Structural Basis of H+-Dependent Conformational Change in a Bacterial MATE Transporter.
Structure, 27, 2019
|
|
3NKP
 
 | | Crystal structure of mouse autotaxin in complex with 18:1-LPA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nishimasu, H, Ishitani, R, Mihara, E, Takagi, J, Aoki, J, Nureki, O. | | Deposit date: | 2010-06-20 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Crystal structure of autotaxin and insight into GPCR activation by lipid mediators
Nat.Struct.Mol.Biol., 18, 2011
|
|
2AGT
 
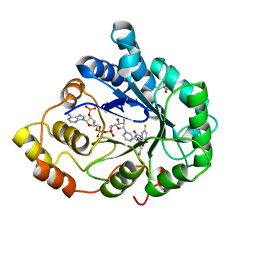 | | Aldose Reductase Mutant Leu 300 Pro complexed with Fidarestat | | Descriptor: | (2S,4S)-2-AMINOFORMYL-6-FLUORO-SPIRO[CHROMAN-4,4'-IMIDAZOLIDINE]-2',5'-DIONE, Aldose reductase, CHLORIDE ION, ... | | Authors: | Petrova, T, Steuber, H, Hazemann, I, Cousido-Siah, A, Mitschler, A, Chung, R, Oka, M, Klebe, G, El-Kabbani, O, Joachimiak, A, Podjarny, A. | | Deposit date: | 2005-07-27 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Factorizing Selectivity Determinants of Inhibitor Binding toward Aldose and Aldehyde Reductases: Structural and Thermodynamic Properties of the Aldose Reductase Mutant Leu300Pro-Fidarestat Complex
J.Med.Chem., 48, 2005
|
|
3NKR
 
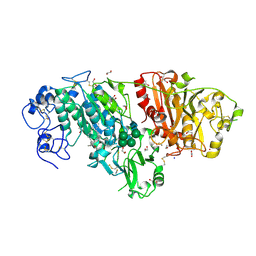 | | Crystal structure of mouse autotaxin in complex with 22:6-LPA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (4Z,7E,10E,13Z,16Z,19Z)-docosa-4,7,10,13,16,19-hexaenoate, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nishimasu, H, Ishitani, R, Mihara, E, Takagi, J, Aoki, J, Nureki, O. | | Deposit date: | 2010-06-20 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.704 Å) | | Cite: | Crystal structure of autotaxin and insight into GPCR activation by lipid mediators
Nat.Struct.Mol.Biol., 18, 2011
|
|
2ASL
 
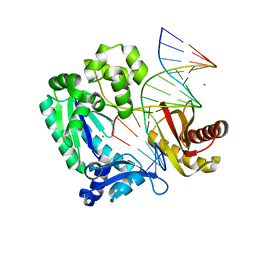 | | oxoG-modified Postinsertion Binary Complex | | Descriptor: | 5'-D(*CP*T*AP*AP*CP*(8OG)P*CP*TP*AP*CP*CP*AP*TP*CP*CP*AP*AP*CP*C)-3', 5'-D(*GP*GP*TP*TP*GP*GP*AP*TP*GP*GP*TP*AP*GP*(DOC))-3', CALCIUM ION, ... | | Authors: | Rechkoblit, O, Malinina, L, Cheng, Y, Kuryavyi, V, Broyde, S, Geacintov, N.E, Patel, D.J. | | Deposit date: | 2005-08-23 | | Release date: | 2006-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Stepwise Translocation of Dpo4 Polymerase during Error-Free Bypass of an oxoG Lesion
Plos Biol., 4, 2006
|
|
3P1A
 
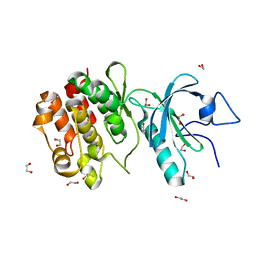 | | Structure of human Membrane-associated Tyrosine- and Threonine-specific cdc2-inhibitory kinase MYT1 (PKMYT1) | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase | | Authors: | Chaikuad, A, Eswaran, J, Fedorov, O, Cooper, C.D.O, Kroeler, T, Vollmar, M, Krojer, T, Berridge, G, Muniz, J.R.C, Pike, A.C.W, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-09-30 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of human Membrane-associated Tyrosine- and Threonine-specific cdc2-inhibitory kinase MYT1 (PKMYT1)
To be Published
|
|
2AVI
 
 | |
6FWG
 
 | | Structure of an E333Q variant of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with tetramannoside yeast mannan fragment | | Descriptor: | ACETATE ION, Glycosyl hydrolase family 71, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-methyl alpha-D-mannopyranoside | | Authors: | Sobala, L.F, Speciale, G, Hakki, Z, Fernandes, P.Z, Raich, L, Rojas-Cervellera, V, Bennet, A, Thompson, A.J, Bernardo-Seisdedos, G, Millet, O, Zhu, S, Lu, D, Sollogoub, M, Rovira, C, Jimenez-Barbero, J, Davies, G.J, Williams, S.J. | | Deposit date: | 2018-03-06 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | An Epoxide Intermediate in Glycosidase Catalysis.
Acs Cent.Sci., 6, 2020
|
|
6FWM
 
 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with alpha-Glc-1,3-1,2-anhydro-mannose hydrolyzed by enzyme | | Descriptor: | (1~{S},2~{R},3~{S},4~{R},5~{R})-5-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, ACETATE ION, Glycosyl hydrolase family 71, ... | | Authors: | Sobala, L.F, Speciale, G, Hakki, Z, Fernandes, P.Z, Raich, L, Rojas-Cervellera, V, Bennet, A, Thompson, A.J, Bernardo-Seisdedos, G, Millet, O, Zhu, S, Lu, D, Sollogoub, M, Rovira, C, Jimenez-Barbero, J, Davies, G.J, Williams, S.J. | | Deposit date: | 2018-03-06 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | An Epoxide Intermediate in Glycosidase Catalysis.
Acs Cent.Sci., 6, 2020
|
|
6FWQ
 
 | | Structure of an E333Q variant of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with alpha-1,3-mannobiose and alpha-1,2-mannobiose | | Descriptor: | Glycosyl hydrolase family 71, SODIUM ION, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose, ... | | Authors: | Sobala, L.F, Speciale, G, Hakki, Z, Fernandes, P.Z, Raich, L, Rojas-Cervellera, V, Bennet, A, Thompson, A.J, Bernardo-Seisdedos, G, Millet, O, Zhu, S, Lu, D, Sollogoub, M, Rovira, C, Jimenez-Barbero, J, Davies, G.J, Williams, S.J. | | Deposit date: | 2018-03-07 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | An Epoxide Intermediate in Glycosidase Catalysis.
Acs Cent.Sci., 6, 2020
|
|
1X96
 
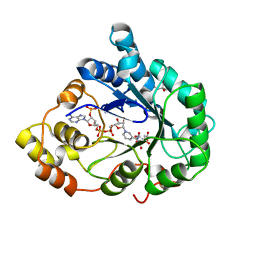 | | Crystal structure of Aldose Reductase with citrates bound in the active site | | Descriptor: | CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, aldose reductase | | Authors: | El-Kabbani, O, Darmanin, C, Oka, M, Schulze-Briese, C, Tomizaki, T, Hazemann, I, Mitschler, A, Podjarny, A. | | Deposit date: | 2004-08-19 | | Release date: | 2004-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-Resolution Structures of Human Aldose Reductase Holoenzyme in Complex with Stereoisomers of the Potent Inhibitor Fidarestat: Stereospecific Interaction between the Enzyme and a Cyclic Imide Type Inhibitor
J.Med.Chem., 47, 2004
|
|
3PES
 
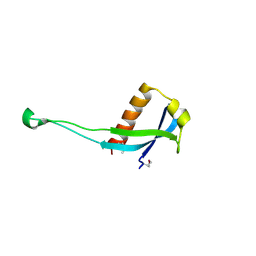 | | Crystal structure of uncharacterized protein from Pseudomonas phage YuA | | Descriptor: | Uncharacterized protein gp49 | | Authors: | Nocek, B, Stein, A, Evdokimove, A, Egorova, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-10-27 | | Release date: | 2010-12-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of uncharacterized protein from Pseudomonas phage YuA
TO BE PUBLISHED
|
|
1X97
 
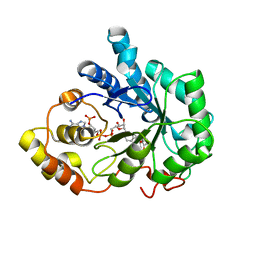 | | Crystal structure of Aldose Reductase complexed with 2R4S (Stereoisomer of Fidarestat, 2S4S) | | Descriptor: | (2R,4S)-2-AMINOFORMYL-6-FLUORO-SPIRO[CHROMAN-4,4'-IMIDAZOLIDINE]-2',5'-DIONE, Aldose Reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | El-Kabbani, O, Darmanin, C, Oka, M, Schulze-Briese, C, Tomizaki, T, Hazemann, I, Mitschler, A, Podjarny, A. | | Deposit date: | 2004-08-19 | | Release date: | 2004-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-Resolution Structures of Human Aldose Reductase Holoenzyme in Complex with Stereoisomers of the Potent Inhibitor Fidarestat: Stereospecific Interaction between the Enzyme and a Cyclic Imide Type Inhibitor
J.Med.Chem., 47, 2004
|
|
1XBW
 
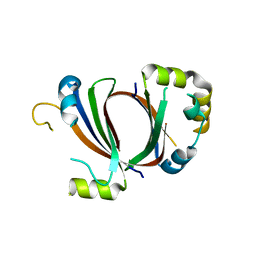 | | 1.9A Crystal Structure of the protein isdG from Staphylococcus aureus aureus, Structural genomics, MCSG | | Descriptor: | hypothetical protein isdG | | Authors: | Zhang, R, Wu, R, Joachimiak, G, Schneewind, O, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-31 | | Release date: | 2004-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Staphylococcus aureus IsdG and IsdI, heme-degrading enzymes with structural similarity to monooxygenases.
J.Biol.Chem., 280, 2005
|
|
6HF6
 
 | | Crystal structure of the Protease 1 (E29A,E60A,E80A) from Pyrococcus horikoshii co-crystallized with Tb-Xo4. | | Descriptor: | Deglycase PH1704, MALONATE ION, TERBIUM(III) ION, ... | | Authors: | Engilberge, S, Wagner, T, Santoni, G, Breyton, C, Shima, S, Franzetti, B, Riobe, F, Maury, O, Girard, E. | | Deposit date: | 2018-08-21 | | Release date: | 2019-06-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein crystal structure determination with the crystallophore, a nucleating and phasing agent.
J.Appl.Crystallogr., 52, 2019
|
|
3PDO
 
 | | Crystal Structure of HLA-DR1 with CLIP102-120 | | Descriptor: | FORMIC ACID, GLYCEROL, HLA class II histocompatibility antigen gamma chain, ... | | Authors: | Gunther, S, Schlundt, A, Sticht, J, Roske, Y, Heinemann, U, Wiesmuller, K.-H, Jung, G, Falk, K, Rotzschke, O, Freund, C. | | Deposit date: | 2010-10-23 | | Release date: | 2010-12-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Bidirectional binding of invariant chain peptides to an MHC class II molecule.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7SL8
 
 | | CryoEM structure of SGLT1 at 3.4 A resolution | | Descriptor: | CHOLESTEROL, Sodium/glucose cotransporter 1, nanobody Nb1 | | Authors: | Qu, Q, Han, L, Panova, O, Feng, L, Skiniotis, G. | | Deposit date: | 2021-10-23 | | Release date: | 2021-12-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure and mechanism of the SGLT family of glucose transporters.
Nature, 601, 2022
|
|
7SL9
 
 | | CryoEM structure of SMCT1 | | Descriptor: | Sodium-coupled monocarboxylate transporter 1, butanoic acid, nanobody Nb2 | | Authors: | Qu, Q, Han, L, Panova, O, Feng, L, Skiniotis, G. | | Deposit date: | 2021-10-23 | | Release date: | 2021-12-15 | | Last modified: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure and mechanism of the SGLT family of glucose transporters.
Nature, 601, 2022
|
|
7SLA
 
 | | CryoEM structure of SGLT1 at 3.15 Angstrom resolution | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Sodium/glucose cotransporter 1, nanobody Nb1 | | Authors: | Qu, Q, Han, L, Panova, O, Feng, L, Skiniotis, G. | | Deposit date: | 2021-10-23 | | Release date: | 2021-12-15 | | Last modified: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structure and mechanism of the SGLT family of glucose transporters.
Nature, 601, 2022
|
|
2ARF
 
 | | Solution structure of the Wilson ATPase N-domain in the presence of ATP | | Descriptor: | WILSON DISEASE ATPASE | | Authors: | Dmitriev, O, Tsivkovskii, R, Abildgaard, F, Morgan, C.T, Markley, J.L, Lutsenko, S. | | Deposit date: | 2005-08-19 | | Release date: | 2006-02-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-domain of Wilson disease protein: Distinct nucleotide-binding environment and effects of disease mutations
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3PFI
 
 | | 2.7 Angstrom resolution crystal structure of a probable holliday junction DNA helicase (ruvB) from Campylobacter jejuni subsp. jejuni NCTC 11168 in complex with adenosine-5'-diphosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase ruvB | | Authors: | Halavaty, A.S, Wawrzak, Z, Skarina, T, Onopriyenko, O, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-10-28 | | Release date: | 2010-11-10 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.695 Å) | | Cite: | 2.7 Angstrom resolution crystal structure of a probable holliday junction DNA helicase (ruvB) from Campylobacter jejuni subsp. jejuni NCTC 11168 in complex with adenosine-5'-diphosphate
To be Published
|
|
3PRY
 
 | | Crystal structure of the middle domain of human HSP90-beta refined at 2.3 A resolution | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Heat shock protein HSP 90-beta, ... | | Authors: | Chaikuad, A, Pilka, E, Sharpe, T.D, Cooper, C.D.O, Phillips, C, Berridge, G, Ayinampudi, V, Fedorov, O, Keates, T, Thangaratnarajah, C, Zimmermann, T, Vollmar, M, Yue, W.W, Che, K.H, Krojer, T, Muniz, J.R.C, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-30 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of the middle domain of human HSP90-beta refined at 2.3 A resolution
To be Published
|
|
2AW5
 
 | | Crystal structure of a human malic enzyme | | Descriptor: | NADP-dependent malic enzyme | | Authors: | Papagrigoriou, E, Berridge, G, Smee, C, Bray, J, Arrowsmith, C, Edwards, A, Weigelt, J, Sundstrom, M, Oppermann, U, Gileadi, O, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-08-31 | | Release date: | 2005-09-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a human malic enzyme
To be published
|
|
