4FCJ
 
 | |
4JZT
 
 | | Crystal structure of the Bacillus subtilis pyrophosphohydrolase BsRppH (E68A mutant) bound to GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, dGTP pyrophosphohydrolase | | Authors: | Piton, J, Larue, V, Thillier, Y, Dorleans, A, Pellegrini, O, Li de la Sierra-Gallay, I, Vasseur, J.J, Debart, F, Tisne, C, Condon, C. | | Deposit date: | 2013-04-03 | | Release date: | 2013-05-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Bacillus subtilis RNA deprotection enzyme RppH recognizes guanosine in the second position of its substrates.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6ZOM
 
 | | Oxidized thioredoxin 1 from the anaerobic bacteria Desulfovibrio vulgaris Hildenborough | | Descriptor: | Thioredoxin | | Authors: | Garcin, E, Bornet, O, Nouailler, M, Pieulle, L, Guerlesquin, F, Sebban-Kreuzer, C. | | Deposit date: | 2020-07-07 | | Release date: | 2021-07-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Glutamate optimizes enzymatic activity under high hydrostatic pressure in Desulfovibrio species: effects on the ubiquitous thioredoxin system.
Extremophiles, 25, 2021
|
|
4JZV
 
 | | Crystal structure of the Bacillus subtilis pyrophosphohydrolase BsRppH bound to a non-hydrolysable triphosphorylated dinucleotide RNA (pcp-pGpG) - second guanosine residue in guanosine binding pocket | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, RNA (5'-R(*(GCP)P*G)-3'), ... | | Authors: | Piton, J, Larue, V, Thillier, Y, Dorleans, A, Pellegrini, O, Li de la Sierra-Gallay, I, Vasseur, J.J, Debart, F, Tisne, C, Condon, C. | | Deposit date: | 2013-04-03 | | Release date: | 2013-05-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bacillus subtilis RNA deprotection enzyme RppH recognizes guanosine in the second position of its substrates.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4GTZ
 
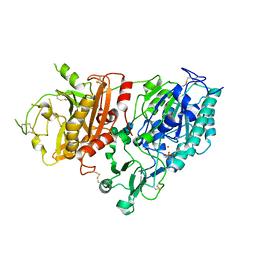 | | Crystal structure of mouse Enpp1 in complex with CMP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Kato, K, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2012-08-29 | | Release date: | 2012-11-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.191 Å) | | Cite: | Crystal structure of Enpp1, an extracellular glycoprotein involved in bone mineralization and insulin signaling.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5JCZ
 
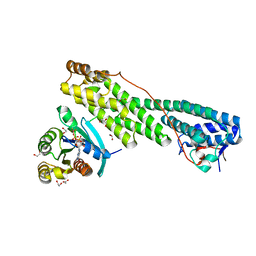 | | Rab11 bound to MyoVa-GTD | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Pylypenko, O, Attanda, W, Gauquelin, C, Malherbes, G, Houdusse, A. | | Deposit date: | 2016-04-15 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.056 Å) | | Cite: | Coordinated recruitment of Spir actin nucleators and myosin V motors to Rab11 vesicle membranes.
Elife, 5, 2016
|
|
5JCY
 
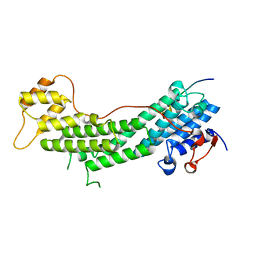 | | Spir2-GTBM bound to MyoVa-GTD | | Descriptor: | Protein spire homolog 2, Unconventional myosin-Va | | Authors: | Pylypenko, O, Malherbes, G, Welz, T, Kerkhoff, E, Houdusse, A. | | Deposit date: | 2016-04-15 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Coordinated recruitment of Spir actin nucleators and myosin V motors to Rab11 vesicle membranes.
Elife, 5, 2016
|
|
4ZNG
 
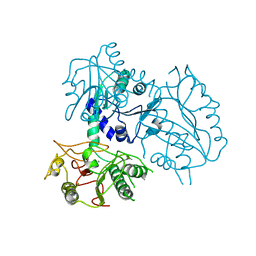 | |
4JZS
 
 | | Crystal structure of the Bacillus subtilis pyrophosphohydrolase BsRppH (E68A mutant) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, dGTP pyrophosphohydrolase | | Authors: | Piton, J, Larue, V, Thillier, Y, Dorleans, A, Pellegrini, O, Li de la Sierra-Gallay, I, Vasseur, J.J, Debart, F, Tisne, C, Condon, C. | | Deposit date: | 2013-04-03 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bacillus subtilis RNA deprotection enzyme RppH recognizes guanosine in the second position of its substrates.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JZU
 
 | | Crystal structure of the Bacillus subtilis pyrophosphohydrolase BsRppH bound to a non-hydrolysable triphosphorylated dinucleotide RNA (pcp-pGpG) - first guanosine residue in guanosine binding pocket | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, RNA (5'-R(*(GCP)P*G)-3'), RNA PYROPHOSPHOHYDROLASE | | Authors: | Piton, J, Larue, V, Thillier, Y, Dorleans, A, Pellegrini, O, Li de la Sierra-Gallay, I, Vasseur, J.J, Debart, F, Tisne, C, Condon, C. | | Deposit date: | 2013-04-03 | | Release date: | 2013-05-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Bacillus subtilis RNA deprotection enzyme RppH recognizes guanosine in the second position of its substrates.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5LH4
 
 | | Trypsin inhibitors for the treatment of pancreatitis - cpd 1 | | Descriptor: | (2~{S},4~{S})-1-[4-(aminomethyl)phenyl]carbonyl-4-(4-cyclopropyl-1,2,3-triazol-1-yl)-~{N}-(2,2-diphenylethyl)pyrrolidine-2-carboxamide, CALCIUM ION, Cationic trypsin, ... | | Authors: | Schiering, N, D'Arcy, A, Skaanderup, P, Simic, O, Brandl, T, Woelcke, J. | | Deposit date: | 2016-07-08 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Trypsin inhibitors for the treatment of pancreatitis.
Bioorg. Med. Chem. Lett., 26, 2016
|
|
5LC9
 
 | |
5LOF
 
 | | Crystal structure of the MBP-MCL1 complex with highly selective and potent inhibitor of MCL1 | | Descriptor: | (2~{R})-2-[5-[3-chloranyl-2-methyl-4-[2-(4-methylpiperazin-1-yl)ethoxy]phenyl]-6-(5-fluoranylfuran-2-yl)thieno[2,3-d]pyrimidin-4-yl]oxy-3-[2-[[2-[2,2,2-tris(fluoranyl)ethyl]pyrazol-3-yl]methoxy]phenyl]propanoic acid, Maltose-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Dokurno, P, Kotschy, A, Szlavik, Z, Murray, J, Davidson, J, Csekei, M, Paczal, A, Szabo, Z, Sipos, S, Radics, G, Proszenyak, A, Balint, B, Ondi, L, Blasko, G, Robertson, A, Surgenor, A, Chen, I, Matassova, N, Smith, J, Pedder, C, Graham, C, Geneste, O. | | Deposit date: | 2016-08-09 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models.
Nature, 538, 2016
|
|
7ADR
 
 | | CO bound as bridging ligand at the active site of vanadium nitrogenase VFe protein | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ... | | Authors: | Rohde, M, Grunau, K, Einsle, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | CO Binding to the FeV Cofactor of CO-Reducing Vanadium Nitrogenase at Atomic Resolution.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5LKA
 
 | | Crystal structure of haloalkane dehalogenase LinB 140A+143L+177W+211L mutant (LinB86) from Sphingobium japonicum UT26 at 1.3 A resolution | | Descriptor: | Haloalkane dehalogenase, THIOCYANATE ION | | Authors: | Degtjarik, O, Rezacova, P, Iermak, I, Chaloupkova, R, Damborsky, J, Kuta Smatanova, I. | | Deposit date: | 2016-07-21 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.298 Å) | | Cite: | Engineering a de novo transport tunnel.
Acs Catalysis, 2016
|
|
5LHM
 
 | | Crystal Structure of SafC from Myxococcus xanthus apo-Form | | Descriptor: | (R,R)-2,3-BUTANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Gerhardt, S, Netzer, J, Einsle, O. | | Deposit date: | 2016-07-12 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Functional and structural characterisation of a bacterial O-methyltransferase and factors determining regioselectivity.
FEBS Lett., 591, 2017
|
|
4Z8F
 
 | |
4Z95
 
 | |
4ZN7
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in complex with Diethylstilbestrol | | Descriptor: | DIETHYLSTILBESTROL, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-05-04 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.934 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5LEF
 
 | | Rab6A:Kif20A complex | | Descriptor: | GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ISOPROPYL ALCOHOL, ... | | Authors: | Bressanelli, G, Pylypenko, O, Houdusse, A. | | Deposit date: | 2016-06-29 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.088 Å) | | Cite: | Coupling fission and exit of RAB6 vesicles at Golgi hotspots through kinesin-myosin interactions.
Nat Commun, 8, 2017
|
|
6I5C
 
 | | Long wavelength native-SAD phasing of Tubulin-Stathmin-TTL complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Basu, S, Olieric, V, Matsugaki, N, Kawano, Y, Takashi, T, Huang, C.Y, Leonarski, F, Yamada, Y, Vera, L, Olieric, N, Basquin, J, Wojdyla, J.A, Diederichs, K, Yamamoto, M, Bunk, O, Wang, M. | | Deposit date: | 2018-11-13 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Long-wavelength native-SAD phasing: opportunities and challenges.
Iucrj, 6, 2019
|
|
5LY6
 
 | | CryoEM structure of the membrane pore complex of Pneumolysin at 4.5A | | Descriptor: | Pneumolysin | | Authors: | van Pee, K, Neuhaus, A, D'Imprima, E, Mills, D.J, Kuehlbrandt, W, Yildiz, O. | | Deposit date: | 2016-09-24 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | CryoEM structures of membrane pore and prepore complex reveal cytolytic mechanism of Pneumolysin.
Elife, 6, 2017
|
|
5LH8
 
 | | Trypsin inhibitors for the treatment of pancreatitis - cpd 8 | | Descriptor: | (2~{S},4~{S})-1-[4-(aminomethyl)-3-methoxy-phenyl]carbonyl-4-(4-cyclopropyl-1,2,3-triazol-1-yl)-~{N}-[(1~{S},2~{R})-2-phenylcyclohexyl]pyrrolidine-2-carboxamide, CALCIUM ION, Cationic trypsin, ... | | Authors: | Schiering, N, D'Arcy, A, Skaanderup, P, Simic, O, Brandl, T, Woelcke, J. | | Deposit date: | 2016-07-08 | | Release date: | 2016-08-10 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Trypsin inhibitors for the treatment of pancreatitis.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
4ZQL
 
 | | Crystal structure of TRIM24 with 3,4-dimethoxy-N-(6-(4-methoxyphenoxy)-1,3-dimethyl-2-oxo-2,3-dihydro-1H-benzo[d]imidazol-5-yl)benzenesulfonamide inhibitor | | Descriptor: | 3,4-dimethoxy-N-[6-(4-methoxyphenoxy)-1,3-dimethyl-2-oxo-2,3-dihydro-1H-benzimidazol-5-yl]benzenesulfonamide, DIMETHYL SULFOXIDE, PENTAETHYLENE GLYCOL, ... | | Authors: | Tallant, C, Structural Genomics Consortium (SGC), Clark, P.G.K, Vieira, L.C.C, Krojer, T, Nunez-Alonso, G, Picaud, S, Fedorov, O, Dixon, D.J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S. | | Deposit date: | 2015-05-10 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of TRIM24 with 3,4-dimethoxy-N-(6-(4-methoxyphenoxy)-1,3-dimethyl-2-oxo-2,3-dihydro-1H-benzo[d]imidazol-5-yl)benzenesulfonamide inhibitor
To Be Published
|
|
4Z0I
 
 | | Crystal structure of a tetramer of GluA2 ligand binding domains bound with glutamate at 1.45 Angstrom resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLUTAMIC ACID, Glutamate receptor 2,Glutamate receptor 2, ... | | Authors: | Baranovic, J, Chebli, M, Salazar, H, Carbone, A.L, Ghisi, V, Faelber, K, Lau, A.Y, Daumke, O, Plested, A.J.R. | | Deposit date: | 2015-03-26 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of the tetrameric wt GluA2 ligand-binding domain bound to glutamate at 1.45 Angstroms resolution
To Be Published
|
|
