195L
 
 | | THERMODYNAMIC AND STRUCTURAL COMPENSATION IN "SIZE-SWITCH" CORE-REPACKING VARIANTS OF T4 LYSOZYME | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, LYSOZYME | | 著者 | Baldwin, E, Xu, J, Hajiseyedjavadi, O, Matthews, B.W. | | 登録日 | 1995-11-06 | | 公開日 | 1996-03-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Thermodynamic and structural compensation in "size-switch" core repacking variants of bacteriophage T4 lysozyme.
J.Mol.Biol., 259, 1996
|
|
1AE4
 
 | |
1APH
 
 | | CONFORMATIONAL CHANGES IN CUBIC INSULIN CRYSTALS IN THE PH RANGE 7-11 | | 分子名称: | 1,2-DICHLOROETHANE, INSULIN A CHAIN (PH 7), INSULIN B CHAIN (PH 7) | | 著者 | Gursky, O, Badger, J, Li, Y, Caspar, D.L.D. | | 登録日 | 1992-10-30 | | 公開日 | 1993-01-15 | | 最終更新日 | 2017-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Conformational changes in cubic insulin crystals in the pH range 7-11.
Biophys.J., 63, 1992
|
|
1B7Y
 
 | | PHENYLALANYL TRNA SYNTHETASE COMPLEXED WITH PHENYLALANINYL-ADENYLATE | | 分子名称: | ADENOSINE-5'-[PHENYLALANINOL-PHOSPHATE], MAGNESIUM ION, PROTEIN (PHENYLALANYL-TRNA SYNTHETASE) | | 著者 | Reshetnikova, L, Moor, N, Lavrik, O, Vassylyev, D.G. | | 登録日 | 1999-01-26 | | 公開日 | 2000-01-26 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structures of phenylalanyl-tRNA synthetase complexed with phenylalanine and a phenylalanyl-adenylate analogue.
J.Mol.Biol., 287, 1999
|
|
198L
 
 | | THERMODYNAMIC AND STRUCTURAL COMPENSATION IN "SIZE-SWITCH" CORE-REPACKING VARIANTS OF T4 LYSOZYME | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, LYSOZYME | | 著者 | Baldwin, E, Xu, J, Hajiseyedjavadi, O, Matthews, B.W. | | 登録日 | 1995-11-06 | | 公開日 | 1996-03-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Thermodynamic and structural compensation in "size-switch" core repacking variants of bacteriophage T4 lysozyme.
J.Mol.Biol., 259, 1996
|
|
196L
 
 | | THERMODYNAMIC AND STRUCTURAL COMPENSATION IN "SIZE-SWITCH" CORE-REPACKING VARIANTS OF T4 LYSOZYME | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, LYSOZYME | | 著者 | Baldwin, E, Xu, J, Hajiseyedjavadi, O, Matthews, B.W. | | 登録日 | 1995-11-06 | | 公開日 | 1996-03-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Thermodynamic and structural compensation in "size-switch" core repacking variants of bacteriophage T4 lysozyme.
J.Mol.Biol., 259, 1996
|
|
3BLM
 
 | |
8DD5
 
 | | Crystal structure of KAT6A in complex with inhibitor CTx-648 (PF-9363) | | 分子名称: | 2,6-dimethoxy-N-{4-methoxy-6-[(1H-pyrazol-1-yl)methyl]-1,2-benzoxazol-3-yl}benzene-1-sulfonamide, Histone acetyltransferase KAT6A, ZINC ION | | 著者 | Greasley, S.E, Johnson, E, Brodsky, O. | | 登録日 | 2022-06-17 | | 公開日 | 2023-07-05 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | Targeting KAT6A/KAT6B dependencies in breast cancer with a novel selective, orally bioavailable KAT6 inhibitor, CTx-648/PF-9363
To Be Published
|
|
7TI6
 
 | |
7VK3
 
 | |
7VK5
 
 | |
7Z74
 
 | | PI3KC2a core in complex with PITCOIN2 | | 分子名称: | 1,2-ETHANEDIOL, Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha, ~{N}-[4-(3-hydroxyphenyl)-1,3-thiazol-2-yl]-2-[4-oxidanylidene-3-(2-phenylethyl)pteridin-2-yl]sulfanyl-ethanamide | | 著者 | Lo, W.T, Roske, Y, Daumke, O, Haucke, V. | | 登録日 | 2022-03-15 | | 公開日 | 2022-08-31 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Development of selective inhibitors of phosphatidylinositol 3-kinase C2 alpha.
Nat.Chem.Biol., 19, 2023
|
|
7Z75
 
 | | PI3KC2a core in complex with PITCOIN3 | | 分子名称: | 1,2-ETHANEDIOL, Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha, SULFATE ION, ... | | 著者 | Lo, W.T, Roske, Y, Daumke, O, Haucke, V. | | 登録日 | 2022-03-15 | | 公開日 | 2022-08-31 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Development of selective inhibitors of phosphatidylinositol 3-kinase C2 alpha.
Nat.Chem.Biol., 19, 2023
|
|
6IS6
 
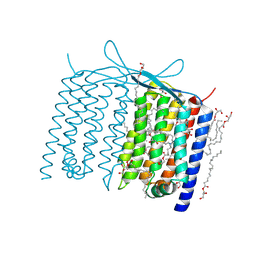 | | Crystal structure of Thermoplasmatales archaeon heliorhodopsin | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, RETINAL, heliorhodopsin | | 著者 | Shihoya, W, Yamashita, K, Nureki, O. | | 登録日 | 2018-11-15 | | 公開日 | 2019-09-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of heliorhodopsin.
Nature, 574, 2019
|
|
1XXM
 
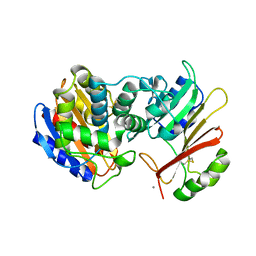 | | The modular architecture of protein-protein binding site | | 分子名称: | Beta-lactamase TEM, Beta-lactamase inhibitory protein, CALCIUM ION | | 著者 | Reichmann, D, Rahat, O, Albeck, S, Meged, R, Dym, O, Schreiber, G, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2004-11-07 | | 公開日 | 2005-01-18 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The modular architecture of protein-protein binding interfaces
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1Y7X
 
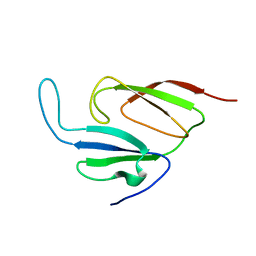 | | Solution structure of a two-repeat fragment of major vault protein | | 分子名称: | Major vault protein | | 著者 | Kozlov, G, Vavelyuk, O, Minailiuc, O, Banville, D, Gehring, K, Ekiel, I. | | 登録日 | 2004-12-10 | | 公開日 | 2005-12-20 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a two-repeat fragment of major vault protein.
J.Mol.Biol., 356, 2006
|
|
1ZE3
 
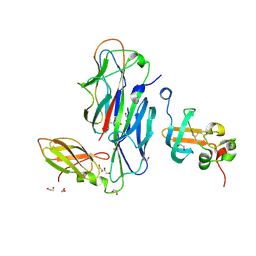 | | Crystal Structure of the Ternary Complex of FIMD (N-Terminal Domain) with FIMC and the Pilin Domain of FIMH | | 分子名称: | 1,2-ETHANEDIOL, Chaperone protein fimC, FimH protein, ... | | 著者 | Nishiyama, M, Horst, R, Eidam, O, Herrmann, T, Ignatov, O, Vetsch, M, Bettendorff, P, Jelesarov, I, Grutter, M.G, Wuthrich, K, Glockshuber, R, Capitani, G. | | 登録日 | 2005-04-17 | | 公開日 | 2005-06-14 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Structural basis of chaperone-subunit complex recognition by the type 1 pilus assembly platform FimD.
Embo J., 24, 2005
|
|
6T33
 
 | | The unusual structure of Ruminococcin C1 antimicrobial peptide confers activity against clinical pathogens | | 分子名称: | Ruminococcin C | | 著者 | Chiumento, S, Roblin, C, Bornet, O, Nouailler, M, Muller, C, Basset, C, Kieffer-Jaquinod, S, Coute, Y, Torelli, S, Le Pape, L, Shunemann, V, Jeannot, K, Nicoletti, C, Iranzo, O, Maresca, M, Giardina, T, Fons, M, Devillard, E, Perrier, J, Atta, M, Guerlesquin, F, Lafond, M, Duarte, V. | | 登録日 | 2019-10-10 | | 公開日 | 2020-08-12 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The unusual structure of Ruminococcin C1 antimicrobial peptide confers clinical properties.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3KVW
 
 | | Crystal Structure of dual-specificity tyrosine phosphorylation regulated kinase 2 (DYRK2) in complex with an indirubin ligand | | 分子名称: | (2Z,3E)-7'-bromo-3-(hydroxyimino)-2'-oxo-1,1',2',3-tetrahydro-2,3'-biindole-5-carboxylic acid, CHLORIDE ION, Dual specificity tyrosine-phosphorylation-regulated kinase 2 | | 著者 | Filippakopoulos, P, Myrianthopoulos, V, Kritsanida, M, Magiatis, P, Skaltsounis, A.L, Soundararajan, M, Krojer, T, Gileadi, O, Hapka, E, Fedorov, O, Berridge, G, Wang, J, Shrestha, L, Vollmar, M, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Bountra, C, Mikros, E, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2009-11-30 | | 公開日 | 2010-01-05 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Crystal Structure of dual-specificity tyrosine phosphorylation regulated kinase 2 (DYRK2) in complex with an indirubin ligand
To be Published
|
|
3LQY
 
 | | Crystal structure of putative isochorismatase hydrolase from Oleispira antarctica | | 分子名称: | GLYCEROL, putative isochorismatase hydrolase | | 著者 | Goral, A, Chruszcz, M, Kagan, O, Cymborowski, M, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-02-10 | | 公開日 | 2010-03-16 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal structure of a putative isochorismatase hydrolase from Oleispira antarctica.
J.Struct.Funct.Genom., 13, 2012
|
|
5LN1
 
 | | STRUCTURE OF UBIQUITYLATED-RPN10 FROM YEAST; | | 分子名称: | 26S proteasome regulatory subunit RPN10, Polyubiquitin-B | | 著者 | Keren-Kaplan, T, Attali, I, Levin-Kravets, O, Prag, G. | | 登録日 | 2016-08-02 | | 公開日 | 2016-10-19 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.14 Å) | | 主引用文献 | Structure of ubiquitylated-Rpn10 provides insight into its autoregulation mechanism.
Nat Commun, 7, 2016
|
|
3NQ8
 
 | | Optimization of the in silico designed Kemp eliminase KE70 by computational design and directed evolution R4 8/5A | | 分子名称: | BENZAMIDINE, NITRATE ION, deoxyribose phosphate aldolase | | 著者 | Khersonsky, O, Rothlisberge, D, Wollacott, A.M, Dym, O, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2010-06-29 | | 公開日 | 2011-02-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Optimization of the in-silico-designed kemp eliminase KE70 by computational design and directed evolution
J.Mol.Biol., 407, 2011
|
|
6FGZ
 
 | |
7AQ0
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, D576A/S550A | | 分子名称: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | 著者 | Zhang, L, Bill, E, Kroneck, P.M.H, Einsle, O. | | 登録日 | 2020-10-20 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.584 Å) | | 主引用文献 | Histidine-Gated Proton-Coupled Electron Transfer to the Cu A Site of Nitrous Oxide Reductase.
J.Am.Chem.Soc., 143, 2021
|
|
1BLH
 
 | |
