4B9P
 
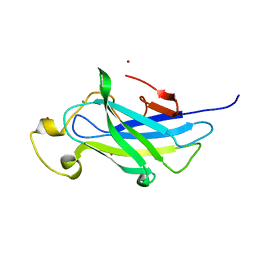 | | Biomass sensoring module from putative Rsgi2 protein of Clostridium thermocellum resemble family 3 carbohydrate-binding module of cellulosome | | Descriptor: | CALCIUM ION, TYPE 3A CELLULOSE-BINDING DOMAIN PROTEIN, ZINC ION | | Authors: | Yaniv, O, Shimon, L.J.W, Bayer, E.A, Lamed, R, Frolow, F. | | Deposit date: | 2012-09-06 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.182 Å) | | Cite: | Fine-Structural Variance of Family 3 Carbohydrate-Binding Modules as Extracellular Biomass-Sensing Components of Clostridium Thermocellum Anti-Sigma(I) Factors.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4BDQ
 
 | | Crystal structure of the GluK2 R775A LBD dimer in complex with glutamate | | Descriptor: | GLUTAMATE RECEPTOR, IONOTROPIC KAINATE 2, GLUTAMIC ACID, ... | | Authors: | Nayeem, N, Mayans, O, Green, T. | | Deposit date: | 2012-10-05 | | Release date: | 2013-04-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Correlating Efficacy and Desensitization with Gluk2 Ligand-Binding Domain Movements.
Open Biol., 3, 2013
|
|
4AVM
 
 | | Crystal structure of the N-BAR domain of human bridging integrator 2. | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BRIDGING INTEGRATOR 2 | | Authors: | Allerston, C.K, Krojer, T, Cooper, C.D.O, Vollmar, M, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Gileadi, O. | | Deposit date: | 2012-05-28 | | Release date: | 2012-06-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal Structure of the N-Bar Domain of Human Bridging Integrator 2.
To be Published
|
|
4BHN
 
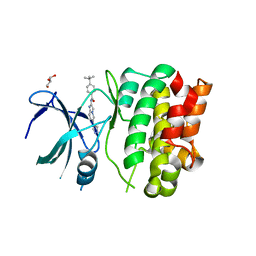 | | Crystal Structures of Ask1-inhibitor Complexes | | Descriptor: | 4-tert-butyl-N-[6-(1H-pyrazol-4-yl)imidazo[1,2-a]pyridin-2-yl]benzamide, GLYCEROL, MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 5 | | Authors: | Singh, O, Shillings, A, Craggs, P, Wall, I, Rowland, P, Skarzynski, T, Hobbs, C.I, Hardwick, P, Tanner, R, Blunt, M, Witty, D.R, Smith, K.J. | | Deposit date: | 2013-04-04 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Ask1-Inhibtor Complexes Provide a Platform for Structure Based Drug Design.
Protein Sci., 22, 2013
|
|
4BIC
 
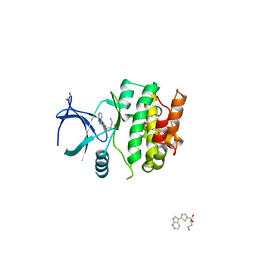 | | Crystal Structures of Ask1-inhibitor Complexes | | Descriptor: | GLYCEROL, MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 5, N-(2-aminoethyl)-5-{1H-pyrrolo[2,3-b]pyridin-3-yl}thiophene-2-sulfonamide | | Authors: | Singh, O, Shillings, A, Craggs, P, Wall, I, Rowland, P, Skarzynski, T, Hobbs, C.I, Hardwick, P, Tanner, R, Blunt, M, Witty, D.R, Smith, K.J. | | Deposit date: | 2013-04-10 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Crystal Structures of Ask1-Inhibtor Complexes Provide a Platform for Structure Based Drug Design.
Protein Sci., 22, 2013
|
|
4BLI
 
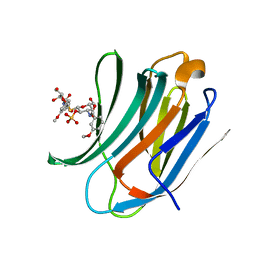 | | Galectin-3c in complex with Bisamido-thiogalactoside derivate 1 | | Descriptor: | (3-Deoxy-3-(3-methoxy-benzamido)-b-D-galactopyranosyl)-(3-deoxy-3-(3-methoxy-benzamido)-2-O-sulfo-b-D-galactopyranosyl)-sulfide, GALECTIN-3 | | Authors: | Noresson, A.L, Oberg, C.T, Engstrom, O, Hakansson, M, Logan, D.T, Leffler, H, Nilsson, U.J. | | Deposit date: | 2013-05-03 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Controlling Protein Conformation Through Electronic Fine-Tuning of Arginine-Arene Interactions: Synthetic, Structural, and Biological Studies
To be Published
|
|
4B60
 
 | | Structure of rFnBPA(189-505) in complex with fibrinogen gamma chain C- terminal peptide | | Descriptor: | CALCIUM ION, FIBRINOGEN GAMMA CHAIN, FIBRONECTIN-BINDING PROTEIN A | | Authors: | Stemberk, V, Moroz, O, Atkin, K.E, Turkenburg, J.P, Potts, J.R. | | Deposit date: | 2012-08-08 | | Release date: | 2013-10-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Evidence for Steric Regulation of Fibrinogen Binding to Staphylococcus Aureus Fibronectin-Binding Protein a (Fnbpa).
J.Biol.Chem., 289, 2014
|
|
4D1U
 
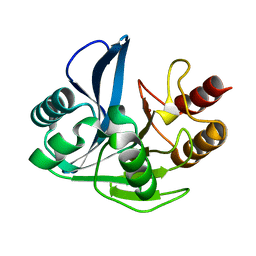 | | A D120A mutant of VIM-7 from Pseudomonas aeruginosa | | Descriptor: | METALLO-B-LACTAMASE, ZINC ION | | Authors: | Leiros, H.-K.S, Skagseth, S, Edvardsen, K.S.W, Lorentzen, M.S, Bjerga, G.E.K, Leiros, I, Samuelsen, O. | | Deposit date: | 2014-05-05 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | His224 Alters the R2 Drug Binding Site and Phe218 Influences the Catalytic Efficiency in the Metallo-Beta-Lactamase Vim-7.
Antimicrob.Agents Chemother., 58, 2014
|
|
4CZY
 
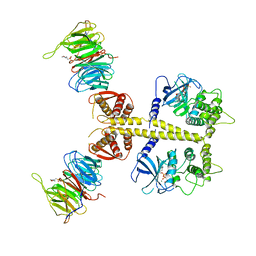 | | Complex of Neurospora crassa PAN2 (WD40-CS1) with PAN3 (pseudokinase and C-term) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, PAB-DEPENDENT POLY(A)-SPECIFIC RIBONUCLEASE SUBUNIT PAN2, ... | | Authors: | Jonas, S, Izaurralde, E, Weichenrieder, O. | | Deposit date: | 2014-04-22 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | An Asymmetric Pan3 Dimer Recruits a Single Pan2 Exonuclease to Mediate Mrna Deadenylation and Decay.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4CZB
 
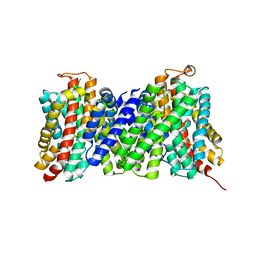 | | Structure of the sodium proton antiporter MjNhaP1 from Methanocaldococcus jannaschii at pH 8. | | Descriptor: | NA(+)/H(+) ANTIPORTER 1, POTASSIUM ION, TRIS(HYDROXYETHYL)AMINOMETHANE | | Authors: | Woehlert, D, Paulino, C, Kapotova, E, Kuhlbrandt, W, Yildiz, O. | | Deposit date: | 2014-04-16 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | 3D Em Map of the Sodium Proton Antiporter Mjnhap1 from Methanocaldococcus Jannaschii
Elife, 3, 2014
|
|
4D0M
 
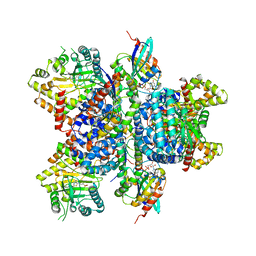 | | Phosphatidylinositol 4-kinase III beta in a complex with Rab11a-GTP- gamma-S and the Rab-binding domain of FIP3 | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, N-(5-(4-CHLORO-3-(2-HYDROXY-ETHYLSULFAMOYL)- PHENYLTHIAZOLE-2-YL)-ACETAMIDE, ... | | Authors: | Burke, J.E, Inglis, A.J, Perisic, O, Masson, G.R, McLaughlin, S.H, Rutaganira, F, Shokat, K.M, Williams, R.L. | | Deposit date: | 2014-04-29 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Structures of Pi4Kiiibeta Complexes Show Simultaneous Recruitment of Rab11 and its Effectors.
Science, 344, 2014
|
|
4D0K
 
 | |
4D1T
 
 | | High resolution structure of native tVIM-7 from Pseudomonas aeruginosa | | Descriptor: | METALLO-B-LACTAMASE, ZINC ION | | Authors: | Leiros, H.-K.S, Skagseth, S, Edvardsen, K.S.W, Lorentzen, M.S, Bjerga, G.E.K, Leiros, I, Samuelsen, O. | | Deposit date: | 2014-05-05 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | His224 Alters the R2 Drug Binding Site and Phe218 Influences the Catalytic Efficiency in the Metallo-Beta-Lactamase Vim-7.
Antimicrob.Agents Chemother., 58, 2014
|
|
4D1V
 
 | | A F218Y mutant of VIM-7 from Pseudomonas aeruginosa | | Descriptor: | METALLO-B-LACTAMASE, ZINC ION | | Authors: | Leiros, H.-K.S, Skagseth, S, Edvardsen, K.S.W, Lorentzen, M.S, Bjerga, G.E.K, Leiros, I, Samuelsen, O. | | Deposit date: | 2014-05-05 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | His224 Alters the R2 Drug Binding Site and Phe218 Influences the Catalytic Efficiency in the Metallo-Beta-Lactamase Vim-7.
Antimicrob.Agents Chemother., 58, 2014
|
|
4CB7
 
 | | Structure of Influenza A H5N1 PB2 cap-binding domain with bound cap analogue (compound 8e) | | Descriptor: | 5-[2-(2-azanyl-7-methyl-6-oxidanylidene-1H-purin-9-ium-9-yl)ethanoyl]-2-oxidanyl-benzoic acid, GLYCEROL, POLYMERASE BASIC SUBUNIT 2 | | Authors: | Pautus, S, Sehr, P, Lewis, J, Fortune, A, Wolkerstorfer, A, Szolar, O, Gulligay, D, Lunardi, T, Decout, J.L, Cusack, S. | | Deposit date: | 2013-10-10 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | New 7-Methyl-Guanosine Derivatives Targeting the Influenza Polymerase Pb2 CAP-Binding Domain
J.Med.Chem., 56, 2013
|
|
4CBK
 
 | | The c-ring ion binding site of the ATP synthase from Bacillus pseudofirmus OF4 is adapted to alkaliphilic cell physiology | | Descriptor: | ATP SYNTHASE SUBUNIT C, DODECYL-BETA-D-MALTOSIDE, SODIUM ION, ... | | Authors: | Preiss, L, Yildiz, O, Meier, T. | | Deposit date: | 2013-10-14 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | The C-Ring Ion-Binding Site of the ATP Synthase from Bacillus Pseudofirmus of4 is Adapted to Alkaliphilic Lifestyle.
Mol.Microbiol., 92, 2014
|
|
4C8Z
 
 | | Cas6 (TTHA0078) product complex | | Descriptor: | CAS6A, POTASSIUM ION, R1 REPEAT RNA CLEAVAGE PRODUCT, ... | | Authors: | Jinek, M, Niewoehner, O, Doudna, J.A. | | Deposit date: | 2013-10-02 | | Release date: | 2013-11-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Evolution of Crispr RNA Recognition and Processing by Cas6 Endonucleases.
Nucleic Acids Res., 42, 2014
|
|
4BLJ
 
 | | Galectin-3c in complex with Bisamido-thiogalactoside derivate 2 | | Descriptor: | Bis-(3-deoxy-3-(3-methoxy-benzamido)-b-D-galactopyranosyl)-sulfide, GALECTIN-3 | | Authors: | Noresson, A.L, Oberg, C.T, Engstrom, O, Hakansson, M, Logan, D.T, Leffler, H, Nilsson, U.J. | | Deposit date: | 2013-05-03 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Controlling Protein Conformation Through Electronic Fine-Tuning of Arginine-Arene Interactions: Synthetic, Structural, and Biological Studies
To be Published
|
|
4AYW
 
 | | STRUCTURE OF THE HUMAN MITOCHONDRIAL ABC TRANSPORTER, ABCB10 (PLATE FORM) | | Descriptor: | ATP-BINDING CASSETTE SUB-FAMILY B MEMBER 10, CHOLESTEROL HEMISUCCINATE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Pike, A.C.W, Shintre, C.A, Li, Q, Kim, J, von Delft, F, Barr, A.J, Das, S, Chaikuad, A, Xia, X, Quigley, A, Dong, Y, Dong, L, Krojer, T, Vollmar, M, Muniz, J.R.C, Bray, J.E, Berridge, G, Chalk, R, Gileadi, O, Burgess-Brown, N, Shrestha, L, Goubin, S, Yang, J, Mahajan, P, Mukhopadhyay, S, Bullock, A.N, Arrowsmith, C.H, Weigelt, J, Bountra, C, Edwards, A.M, Carpenter, E.P. | | Deposit date: | 2012-06-22 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of Abcb10, a Human ATP-Binding Cassette Transporter in Apo- and Nucleotide-Bound States
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4B96
 
 | | Family 3b carbohydrate-binding module from the biomass sensoring system of Clostridium clariflavum | | Descriptor: | CALCIUM ION, CELLULOSE BINDING DOMAIN-CONTAINING PROTEIN, CHLORIDE ION | | Authors: | Yaniv, O, Reddy, Y.H.K, Yoffe, H, Shimon, L.J.W, Bayer, E.A, Lamed, R, Frolow, F. | | Deposit date: | 2012-09-02 | | Release date: | 2013-09-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.911 Å) | | Cite: | Structure of Cbm3B from the Biomass Sensoring System of Clostridium Clarifalvum
To be Published
|
|
4BBQ
 
 | | Crystal structure of the CXXC and PHD domain of Human Lysine-specific Demethylase 2A (KDM2A)(FBXL11) | | Descriptor: | 1,2-ETHANEDIOL, LYSINE-SPECIFIC DEMETHYLASE 2A, ZINC ION | | Authors: | Allerston, C.K, Watson, A.A, Edlich, C, Li, B, Chen, Y, Ball, L, Krojer, T, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Laue, E.D, Gileadi, O. | | Deposit date: | 2012-09-27 | | Release date: | 2012-10-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal Structure of the Cxxc and Phd Domain of Human Lysine-Specific Demethylase 2A (Kdm2A)(Fbxl11)
To be Published
|
|
4D49
 
 | | Crystal structure of computationally designed armadillo repeat proteins for modular peptide recognition. | | Descriptor: | ARGININE, ARMADILLO REPEAT PROTEIN ARM00027, POLY ARG DECAPEPTIDE | | Authors: | Reichen, C, Forzani, C, Zhou, T, Parmeggiani, F, Fleishman, S.J, Mittl, P.R.E, Madhurantakam, C, Honegger, A, Ewald, C, Zerbe, O, Baker, D, Caflisch, A, Pluckthun, A. | | Deposit date: | 2014-10-27 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Computationally Designed Armadillo Repeat Proteins for Modular Peptide Recognition.
J.Mol.Biol., 428, 2016
|
|
4D4E
 
 | | Crystal structure of computationally designed armadillo repeat proteins for modular peptide recognition. | | Descriptor: | ARMADILLO REPEAT PROTEIN ARM00016, GLYCEROL | | Authors: | Reichen, C, Forzani, C, Zhou, T, Parmeggiani, F, Fleishman, S.J, Mittl, P.R.E, Madhurantakam, C, Honegger, A, Ewald, C, Zerbe, O, Baker, D, Caflisch, A, Pluckthun, A. | | Deposit date: | 2014-10-28 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computationally Designed Armadillo Repeat Proteins for Modular Peptide Recognition.
J.Mol.Biol., 428, 2016
|
|
4DBR
 
 | | Myosin VI D179Y (MD) pre-powerstroke state | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Pylypenko, O, Sweeney, H.L, Houdusse, A. | | Deposit date: | 2012-01-16 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mutations in myosin VI that cause a loss of coordination between heads provide insights into the structural changes underlying force generation and the importance of gating
To be Published
|
|
4E0B
 
 | | 2.17 Angstrom resolution crystal structure of malate dehydrogenase from Vibrio vulnificus CMCP6 | | Descriptor: | ACETATE ION, Malate dehydrogenase | | Authors: | Halavaty, A.S, Wawrzak, Z, Onopriyenko, O, Kwon, K, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-02 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | 2.17 Angstrom resolution crystal structure of malate dehydrogenase from Vibrio vulnificus CMCP6
To be Published
|
|
