5MTK
 
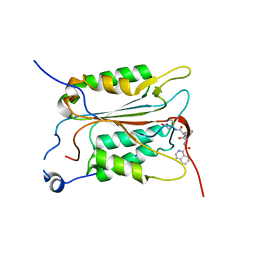 | | Crystal structure of human Caspase-1 with (3S,6S,10aS)-N-((2S,3S)-2-hydroxy-5-oxotetrahydrofuran-3-yl)-6-(isoquinoline-1-carboxamido)-5-oxodecahydropyrrolo[1,2-a]azocine-3-carboxamide (PGE-3935199) | | Descriptor: | (3~{S})-3-[[(3~{S},6~{S},10~{a}~{S})-6-(isoquinolin-1-ylcarbonylamino)-5-oxidanylidene-2,3,6,7,8,9,10,10~{a}-octahydro-1~{H}-pyrrolo[1,2-a]azocin-3-yl]carbonylamino]-4-oxidanyl-butanoic acid, Caspase-1 | | Authors: | Brethon, A, Chantalat, L, Christin, O, Clary, L, Fournier, J.F, Gastreich, M, Harris, C, Pascau, J, Isabet, T, Rodeschin, V, Thoreau, E, Roche, D. | | Deposit date: | 2017-01-09 | | Release date: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Playing against the odds: scaffold hopping from 3D-fragments
To Be Published
|
|
5MVF
 
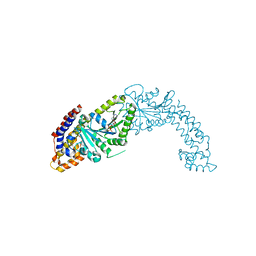 | | Active structure of EHD4 complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, EH domain-containing protein 4, MAGNESIUM ION | | Authors: | Melo, A.A, Daumke, O. | | Deposit date: | 2017-01-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.268 Å) | | Cite: | Structural insights into the activation mechanism of dynamin-like EHD ATPases.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4GEL
 
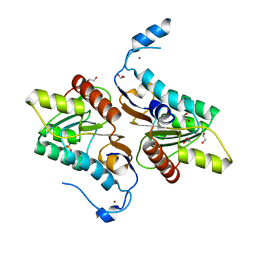 | | Crystal structure of Zucchini | | Descriptor: | 1,2-ETHANEDIOL, Mitochondrial cardiolipin hydrolase, PHOSPHATE ION, ... | | Authors: | Nishimasu, H, Fukuhara, S, Ishitani, R, Nureki, O. | | Deposit date: | 2012-08-02 | | Release date: | 2012-10-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.756 Å) | | Cite: | Structure and function of Zucchini endoribonuclease in piRNA biogenesis
Nature, 491, 2012
|
|
5MEL
 
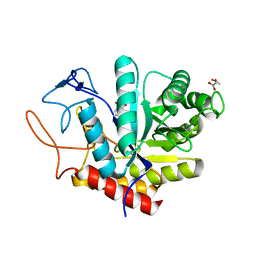 | | Structure of an E333Q variant of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with Glc-alpha-1,3-(3R,4R,5R)-5-(hydroxymethyl)cyclohex-1,2-ene-3,4-diol | | Descriptor: | (1~{R},2~{R},6~{R})-6-(hydroxymethyl)cyclohex-3-ene-1,2-diol, ACETATE ION, Glycosyl hydrolase family 71, ... | | Authors: | Petricevic, M, Sobala, L.F, Fernandes, P.Z, Raich, L, Thompson, A.J, Bernardo-Seisdedos, G, Millet, O, Zhu, S, Sollogoub, M, Rovira, C, Jimenez-Barbero, J, Davies, G.J, Williams, S.J. | | Deposit date: | 2016-11-15 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Contribution of Shape and Charge to the Inhibition of a Family GH99 endo-alpha-1,2-Mannanase.
J. Am. Chem. Soc., 139, 2017
|
|
5MGY
 
 | | Crystal structure of Pseudomonas stutzeri flavinyl transferase ApbE, apo form | | Descriptor: | FAD:protein FMN transferase, MAGNESIUM ION | | Authors: | Zhang, L, Trncik, C, Andrade, S.L.A, Einsle, O. | | Deposit date: | 2016-11-22 | | Release date: | 2016-12-14 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The flavinyl transferase ApbE of Pseudomonas stutzeri matures the NosR protein required for nitrous oxide reduction.
Biochim. Biophys. Acta, 1858, 2016
|
|
5M7F
 
 | | Human porphobilinogen deaminase in complex with DPM cofactor | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, Porphobilinogen deaminase, SULFATE ION | | Authors: | Pluta, P, Millet, O, Roversi, P, Rojas, A.L, Gu, S. | | Deposit date: | 2016-10-27 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structural basis of pyrrole polymerization in human porphobilinogen deaminase.
Biochim Biophys Acta Gen Subj, 1862, 2018
|
|
4GAH
 
 | | Human acyl-CoA thioesterases 4 in complex with undecan-2-one-CoA inhibitor | | Descriptor: | Thioesterase superfamily member 4, [[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(3R)-2,2-dimethyl-3-oxidanyl-4-oxidanylidene-4-[[3-oxidanylidene-3-[2-[(2R)-2-oxidanylundecyl]sulfanylethylamino]propyl]amino]butyl] hydrogen phosphate | | Authors: | Lim, K, Pathak, M.C, Herzberg, O. | | Deposit date: | 2012-07-25 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Correlation of structure and function in the human hotdog-fold enzyme hTHEM4.
Biochemistry, 51, 2012
|
|
5MGP
 
 | | Structural basis for ArfA-RF2 mediated translation termination on stop-codon lacking mRNAs | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Huter, P, Mueller, C, Beckert, B, Arenz, S, Berninghausen, O, Beckmann, R, Wilson, N.D. | | Deposit date: | 2016-11-21 | | Release date: | 2016-12-14 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for ArfA-RF2-mediated translation termination on mRNAs lacking stop codons.
Nature, 541, 2017
|
|
5MOG
 
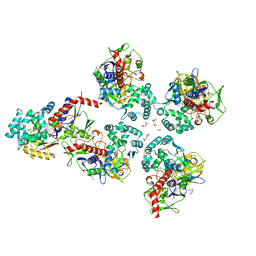 | | Oryza sativa phytoene desaturase inhibited by norflurazon | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Brausemann, A, Gemmecker, S, Koschmieder, J, Beyer, P, Einsle, O. | | Deposit date: | 2016-12-14 | | Release date: | 2017-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structure of Phytoene Desaturase Provides Insights into Herbicide Binding and Reaction Mechanisms Involved in Carotene Desaturation.
Structure, 25, 2017
|
|
1C8T
 
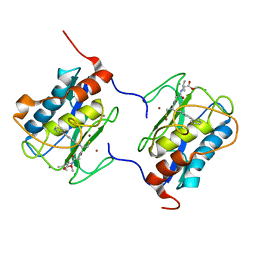 | | HUMAN STROMELYSIN-1 (E202Q) CATALYTIC DOMAIN COMPLEXED WITH RO-26-2812 | | Descriptor: | 2-(2-{2-[(BIPHENYL-4-YLMETHYL)-AMINO]-3-MERCAPTO-PENTANOYLAMINO}-ACETYLAMINO)-3-METHYL-BUTYRIC ACID METHYL ESTER, CALCIUM ION, STROMELYSIN-1, ... | | Authors: | Steele, D.L, el-Kabbani, O, Dunten, P, Crowther, R.L. | | Deposit date: | 1999-07-29 | | Release date: | 2000-07-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Expression, characterization and structure determination of an active site mutant (Glu202-Gln) of mini-stromelysin-1.
Protein Eng., 13, 2000
|
|
5MR8
 
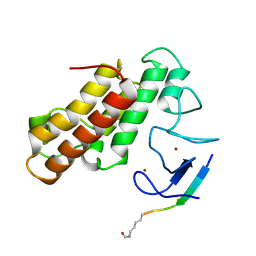 | | Crystal structure of TRIM33 PHD-Bromodomain isoform B in complex with H3K9ac histone peptide | | Descriptor: | E3 ubiquitin-protein ligase TRIM33, Histone H3, ZINC ION | | Authors: | Tallant, C, Savitsky, P, Fedorov, O, Nunez-Alonso, G, Siejka, P, Krojer, T, Williams, E, Srikannathasan, V, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Muller, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-12-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of TRIM33 PHD-Bromodomain isoform B in complex with H3K9ac histone peptide
To Be Published
|
|
5MTN
 
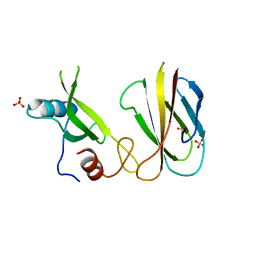 | | Monobody Mb(Lck_1) bound to Lck-Sh2 | | Descriptor: | Monobody Mb(Lck_1), SULFATE ION, Tyrosine-protein kinase Lck | | Authors: | Pojer, F, Kukenshoner, T, Koide, S, Hantschel, O. | | Deposit date: | 2017-01-10 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Selective Targeting of SH2 Domain-Phosphotyrosine Interactions of Src Family Tyrosine Kinases with Monobodies.
J. Mol. Biol., 429, 2017
|
|
6YAF
 
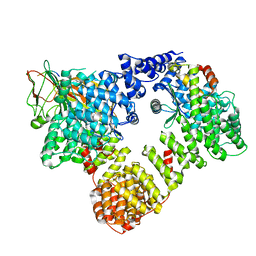 | | AP2 on a membrane containing tyrosine-based cargo peptide | | Descriptor: | AP-2 complex subunit alpha-2, AP-2 complex subunit beta, AP-2 complex subunit mu, ... | | Authors: | Kovtun, O, Kane Dickson, V, Kelly, B.T, Owen, D, Briggs, J.A.G. | | Deposit date: | 2020-03-12 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | Architecture of the AP2/clathrin coat on the membranes of clathrin-coated vesicles.
Sci Adv, 6, 2020
|
|
4FWW
 
 | |
6YE3
 
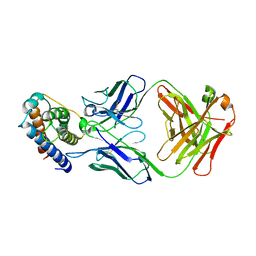 | | IL-2 in complex with a Fab fragment from UFKA-20 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chains: A,D,G, Chains: B,E,H, ... | | Authors: | Karakus, U, Mittl, P, Boyman, O. | | Deposit date: | 2020-03-23 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Receptor-gated IL-2 delivery by an anti-human IL-2 antibody activates regulatory T cells in three different species.
Sci Transl Med, 12, 2020
|
|
1DS5
 
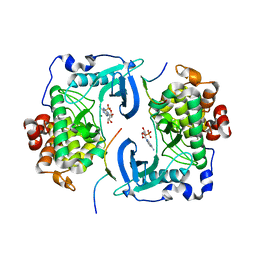 | | DIMERIC CRYSTAL STRUCTURE OF THE ALPHA SUBUNIT IN COMPLEX WITH TWO BETA PEPTIDES MIMICKING THE ARCHITECTURE OF THE TETRAMERIC PROTEIN KINASE CK2 HOLOENZYME. | | Descriptor: | ADENOSINE MONOPHOSPHATE, CASEIN KINASE, ALPHA CHAIN, ... | | Authors: | Battistutta, R, Sarno, S, De Moliner, E, Marin, O, Zanotti, G, Pinna, L.A. | | Deposit date: | 2000-01-07 | | Release date: | 2001-01-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | The crystal structure of the complex of Zea mays alpha subunit with a fragment of human beta subunit provides the clue to the architecture of protein kinase CK2 holoenzyme.
Eur.J.Biochem., 267, 2000
|
|
4DB3
 
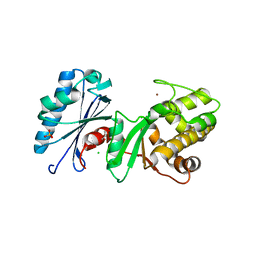 | | 1.95 Angstrom Resolution Crystal Structure of N-acetyl-D-glucosamine kinase from Vibrio vulnificus. | | Descriptor: | CHLORIDE ION, GLYCEROL, N-acetyl-D-glucosamine kinase, ... | | Authors: | Minasov, G, Wawrzak, Z, Onopriyenko, O, Skarina, T, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-01-13 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Resolution Crystal Structure of N-acetyl-D-glucosamine kinase from Vibrio vulnificus.
TO BE PUBLISHED
|
|
1DN6
 
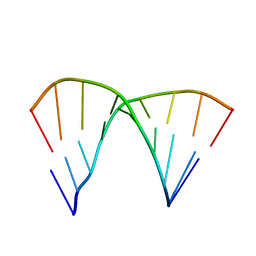 | | THE CRYSTAL STRUCTURE OF D(GGATGGGAG). AN ESSENTIAL PART OF THE BINDING SITE FOR TRANSCRIPTION FACTOR IIIA | | Descriptor: | DNA (5'-D(*CP*TP*CP*CP*CP*AP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*GP*GP*GP*AP*G)-3') | | Authors: | McCall, M, Brown, T, Hunter, W.N, Kennard, O. | | Deposit date: | 1987-05-11 | | Release date: | 1988-01-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of d(GGATGGGAG): an essential part of the binding site for transcription factor IIIA.
Nature, 322, 1986
|
|
6Y77
 
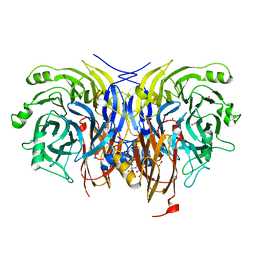 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H326A | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DINUCLEAR COPPER ION, FORMIC ACID, ... | | Authors: | Zhang, L, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-02-28 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A [3Cu:2S] cluster provides insight into the assembly and function of the Cu Z site of nitrous oxide reductase.
Chem Sci, 12, 2021
|
|
6Y57
 
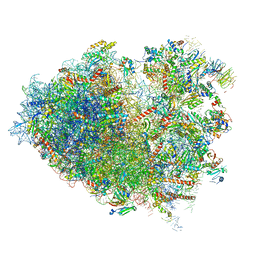 | | Structure of human ribosome in hybrid-PRE state | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Bhaskar, V, Schenk, A.D, Cavadini, S, von Loeffelholz, O, Natchiar, S.K, Klaholz, B.P, Chao, J.A. | | Deposit date: | 2020-02-25 | | Release date: | 2020-04-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Dynamics of uS19 C-Terminal Tail during the Translation Elongation Cycle in Human Ribosomes.
Cell Rep, 31, 2020
|
|
6YGC
 
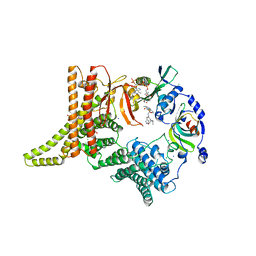 | | Crystal structure of the NatC complex bound to Arl3 peptide and CoA | | Descriptor: | ADP-ribosylation factor-like protein 3, CHLORIDE ION, COENZYME A, ... | | Authors: | Grunwald, S, Hopf, L, Bock-Bierbaum, T, Lally, C.C, Spahn, C.M.T, Daumke, O. | | Deposit date: | 2020-03-27 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.994 Å) | | Cite: | Divergent architecture of the heterotrimeric NatC complex explains N-terminal acetylation of cognate substrates.
Nat Commun, 11, 2020
|
|
6YAI
 
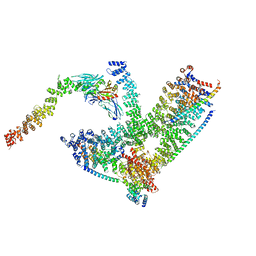 | | Clathrin with bound beta2 appendage of AP2 | | Descriptor: | AP-2 complex subunit beta, Clathrin heavy chain, Clathrin light chain | | Authors: | Kovtun, O, Kane Dickson, V, Kelly, B.T, Owen, D, Briggs, J.A.G. | | Deposit date: | 2020-03-12 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (9.2 Å) | | Cite: | Architecture of the AP2/clathrin coat on the membranes of clathrin-coated vesicles.
Sci Adv, 6, 2020
|
|
5MMV
 
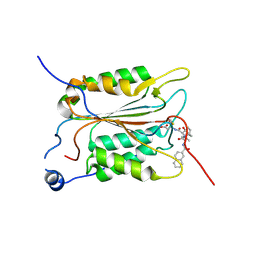 | | Crystal structure of human Caspase-1 with 2-((2-naphthoyl)-L-valyl)-4-hydroxy-N-((3S)-2-hydroxy-5-oxotetrahydrofuran-3-yl)-2-azabicyclo[2.2.2]octane-3-carboxamide (Compound 1) | | Descriptor: | (3~{S})-3-[[(3~{S})-2-[(2~{S})-3-methyl-2-(naphthalen-2-ylcarbonylamino)butanoyl]-4-oxidanyl-2-azabicyclo[2.2.2]octan-3-yl]carbonylamino]-4-oxidanyl-butanoic acid, Caspase-1 | | Authors: | Brethon, A, Chantalat, L, Christin, O, Clary, L, Fournier, J.F, Gastreich, M, Harris, C, Pascau, J, Isabet, T, Rodeschin, V, Thoreau, E, Roche, D. | | Deposit date: | 2016-12-12 | | Release date: | 2017-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of human Caspase-1 with 2-((2-naphthoyl)-L-valyl)-4-hydroxy-N-((3S)-2-hydroxy-5-oxotetrahydrofuran-3-yl)-2-azabicyclo[2.2.2]octane-3-carboxamide (Compound 1)
To Be Published
|
|
6YLN
 
 | | mTurquoise2 SG P212121 - Directional optical properties of fluorescent proteins | | Descriptor: | EGFP, POTASSIUM ION | | Authors: | Myskova, J, Rybakova, O, Brynda, J, Lazar, J. | | Deposit date: | 2020-04-07 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Directionality of light absorption and emission in representative fluorescent proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5MTJ
 
 | | Yes1-SH2 in complex with monobody Mb(Yes_1) | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Monobody Mb(Yes_1), SULFATE ION, ... | | Authors: | Sha, F, Kukenshoner, T, Koide, S, Hantschel, O. | | Deposit date: | 2017-01-09 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Selective Targeting of SH2 Domain-Phosphotyrosine Interactions of Src Family Tyrosine Kinases with Monobodies.
J. Mol. Biol., 429, 2017
|
|
