7ZFN
 
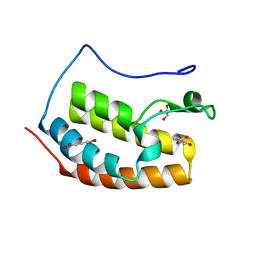 | | BRD4 in complex with FragLite24 | | Descriptor: | 4-bromanyl-2-oxidanyl-benzoic acid, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-04-01 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7ZEF
 
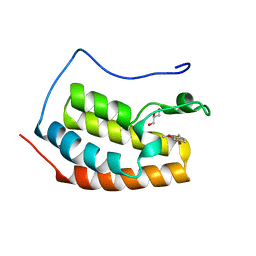 | | BRD4 in complex with FragLite22 | | Descriptor: | (4-bromo-2-methoxyphenyl)methanol, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-03-31 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7ZFT
 
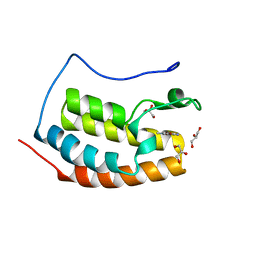 | | BRD4 in complex with FragLite33 | | Descriptor: | 3-azanyl-5-bromanyl-1-methyl-pyridin-2-one, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-04-01 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7ZG1
 
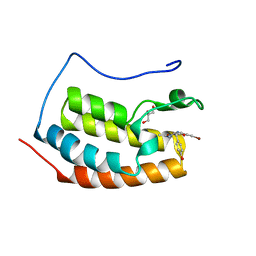 | | BRD4 in complex with PepLite-Tyr | | Descriptor: | Bromodomain-containing protein 4, GLYCEROL, Nalpha-acetyl-N-(3-bromoprop-2-yn-1-yl)-L-tyrosinamide | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-04-01 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7ZG2
 
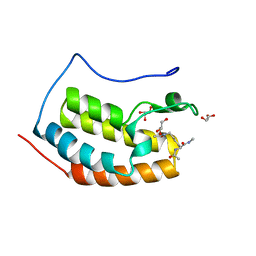 | | BRD4 in complex with Acetyl-Lys | | Descriptor: | (2S)-2,6-diacetamido-N-methyl-hexanamide, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-04-01 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
4B0Z
 
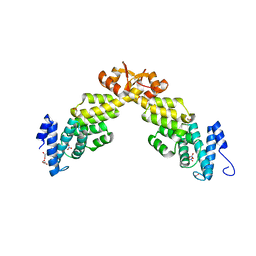 | | Crystal structure of S. pombe Rpn12 | | Descriptor: | 26S PROTEASOME REGULATORY SUBUNIT RPN12, GLYCEROL, MONOTHIOGLYCEROL, ... | | Authors: | Boehringer, J, Riedinger, C, Paraskevopoulos, K, Johnson, E.O.D, Lowe, E.D, Khoudian, C, Smith, D, Noble, M.E.M, Gordon, C, Endicott, J.A. | | Deposit date: | 2012-07-06 | | Release date: | 2012-09-12 | | Last modified: | 2012-11-07 | | Method: | X-RAY DIFFRACTION (1.585 Å) | | Cite: | Structural and Functional Characterisation of Rpn12 Identifies Residues Required for Rpn10 Proteasome Incorporation.
Biochem.J., 448, 2012
|
|
2G9X
 
 | |
5OFM
 
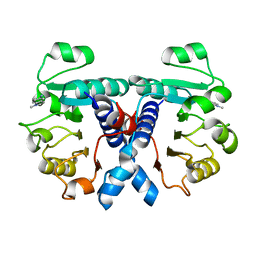 | | Crystal structure of human 3-phosphoglycerate dehydrogenase in complex with 5-amino-1-methyl-1H-indole | | Descriptor: | 1-methylindol-5-amine, D-3-phosphoglycerate dehydrogenase | | Authors: | Unterlass, J.E, Basle, A, Blackburn, T.J, Tucker, J, Cano, C, Noble, M.E.M, Curtin, N.J. | | Deposit date: | 2017-07-11 | | Release date: | 2017-08-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of human 3-phosphoglycerate dehydrogenase in complex with 5-amino-1-methyl-1H-indole
To be published
|
|
2GPN
 
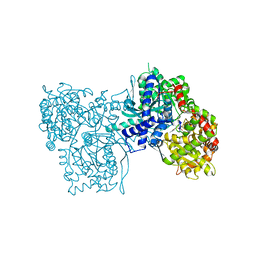 | | 100 K STRUCTURE OF GLYCOGEN PHOSPHORYLASE AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | GLYCOGEN PHOSPHORYLASE B | | Authors: | Gregoriou, M, Noble, M.E.M, Watson, K.A, Garman, E.F, Krulle, T.M, De La Fuente, C, Fleet, G.W.J, Oikonomakos, N.G, Johnson, L.N. | | Deposit date: | 1998-03-26 | | Release date: | 1998-07-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The structure of a glycogen phosphorylase glucopyranose spirohydantoin complex at 1.8 A resolution and 100 K: the role of the water structure and its contribution to binding.
Protein Sci., 7, 1998
|
|
1PHK
 
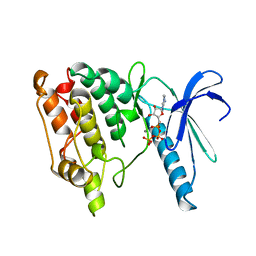 | | TWO STRUCTURES OF THE CATALYTIC DOMAIN OF PHOSPHORYLASE, KINASE: AN ACTIVE PROTEIN KINASE COMPLEXED WITH NUCLEOTIDE, SUBSTRATE-ANALOGUE AND PRODUCT | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, PHOSPHORYLASE KINASE | | Authors: | Owen, D.J, Noble, M.E.M, Garman, E.F, Papageorgiou, A.C, Johnson, L.N. | | Deposit date: | 1996-03-15 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Two structures of the catalytic domain of phosphorylase kinase: an active protein kinase complexed with substrate analogue and product.
Structure, 3, 1995
|
|
2VZG
 
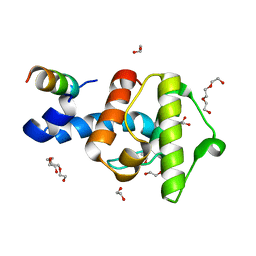 | | Crystal structure of the C-terminal calponin homology domain of alpha- parvin in complex with paxillin LD2 motif | | Descriptor: | 1,2-ETHANEDIOL, Alpha-parvin, Paxillin, ... | | Authors: | Lorenz, S, Vakonakis, I, Lowe, E.D, Campbell, I.D, Noble, M.E.M, Hoellerer, M.K. | | Deposit date: | 2008-08-01 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of the interactions between paxillin LD motifs and alpha-parvin.
Structure, 16, 2008
|
|
2VZI
 
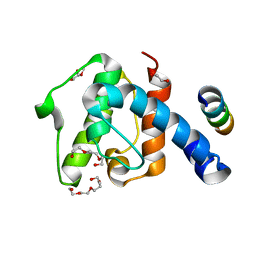 | | Crystal structure of the C-terminal calponin homology domain of alpha- parvin in complex with paxillin LD4 motif | | Descriptor: | 1,2-ETHANEDIOL, Alpha-parvin, Paxillin,Paxillin, ... | | Authors: | Lorenz, S, Vakonakis, I, Lowe, E.D, Campbell, I.D, Noble, M.E.M, Hoellerer, M.K. | | Deposit date: | 2008-08-01 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of the interactions between paxillin LD motifs and alpha-parvin.
Structure, 16, 2008
|
|
2X5N
 
 | | Crystal Structure of the SpRpn10 VWA domain | | Descriptor: | 26S PROTEASOME REGULATORY SUBUNIT RPN10, SULFATE ION | | Authors: | Riedinger, C, Boehringer, J, Trempe, J.-F, Lowe, E.D, Brown, N.R, Gehring, K, Noble, M.E.M, Gordon, C, Endicott, J.A. | | Deposit date: | 2010-02-10 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Structure of Rpn10 and its Interactions with Polyubiquitin Chains and the Proteasome Subunit Rpn12.
J.Biol.Chem., 285, 2010
|
|
2IW8
 
 | | STRUCTURE OF HUMAN THR160-PHOSPHO CDK2-CYCLIN A F82H-L83V-H84D MUTANT WITH AN O6-CYCLOHEXYLMETHYLGUANINE INHIBITOR | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN-A2, MONOTHIOGLYCEROL, ... | | Authors: | Pratt, D.J, Bentley, J, Jewsbury, P, Boyle, F.T, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2006-06-27 | | Release date: | 2006-09-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dissecting the Determinants of Cyclin-Dependent Kinase 2 and Cyclin-Dependent Kinase 4 Inhibitor Selectivity.
J.Med.Chem., 49, 2006
|
|
2IW9
 
 | | STRUCTURE OF HUMAN THR160-PHOSPHO CDK2-CYCLIN A COMPLEXED WITH A BISANILINOPYRIMIDINE INHIBITOR | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN-A2, MAGNESIUM ION, ... | | Authors: | Pratt, D.J, Bentley, J, Jewsbury, P, Boyle, F.T, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2006-06-27 | | Release date: | 2006-09-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dissecting the Determinants of Cyclin-Dependent Kinase 2 and Cyclin-Dependent Kinase 4 Inhibitor Selectivity.
J.Med.Chem., 49, 2006
|
|
2IW6
 
 | | STRUCTURE OF HUMAN THR160-PHOSPHO CDK2-CYCLIN A COMPLEXED WITH A BISANILINOPYRIMIDINE INHIBITOR | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN-A2, MAGNESIUM ION, ... | | Authors: | Pratt, D.J, Bentley, J, Jewsbury, P, Boyle, F.T, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2006-06-26 | | Release date: | 2006-09-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dissecting the Determinants of Cyclin-Dependent Kinase 2 and Cyclin-Dependent Kinase 4 Inhibitor Selectivity.
J.Med.Chem., 49, 2006
|
|
2JCQ
 
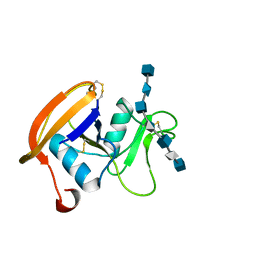 | | The hyaluronan binding domain of murine CD44 in a Type A complex with an HA 8-mer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD44 ANTIGEN, GLYCEROL | | Authors: | Banerji, S, Wright, A.J, Noble, M.E.M, Mahoney, D.J, Campbell, I.D, Day, A.J, Jackson, D.G. | | Deposit date: | 2007-01-03 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structures of the Cd44-Hyaluronan Complex Provide Insight Into a Fundamental Carbohydrate-Protein Interaction.
Nat.Struct.Mol.Biol., 14, 2008
|
|
2JCP
 
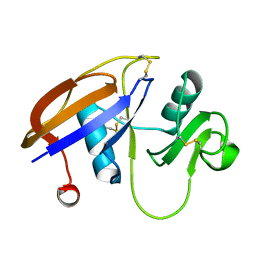 | | The hyaluronan binding domain of murine CD44 | | Descriptor: | CD44 ANTIGEN | | Authors: | Banerji, S, Wright, A.J, Noble, M.E.M, Mahoney, D.J, Campbell, I.D, Day, A.J, Jackson, D.G. | | Deposit date: | 2007-01-03 | | Release date: | 2007-01-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of the Cd44-Hyaluronan Complex Provide Insight Into a Fundamental Carbohydrate-Protein Interaction.
Nat.Struct.Mol.Biol., 14, 2008
|
|
2JCR
 
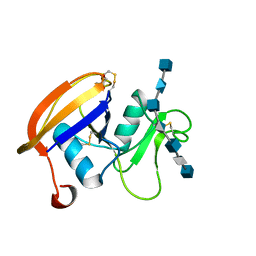 | | The hyaluronan binding domain of murine CD44 in a Type B complex with an HA 8-mer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD44 ANTIGEN, GLYCEROL | | Authors: | Banerji, S, Wright, A.J, Noble, M.E.M, Mahoney, D.J, Campbell, I.D, Day, A.J, Jackson, D.G. | | Deposit date: | 2007-01-03 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the Cd44-Hyaluronan Complex Provide Insight Into a Fundamental Carbohydrate-Protein Interaction.
Nat.Struct.Mol.Biol., 14, 2008
|
|
1OW6
 
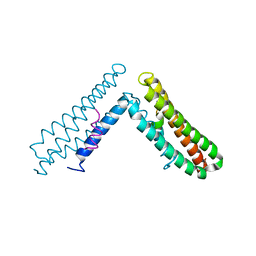 | | Paxillin LD4 motif bound to the Focal Adhesion Targeting (FAT) domain of the Focal Adhesion Kinase | | Descriptor: | Focal adhesion kinase 1, Paxillin | | Authors: | Hoellerer, M.K, Noble, M.E.M, Labesse, G, Campbell, I.D, Werner, J.M, Arold, S.T. | | Deposit date: | 2003-03-28 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular Recognition of Paxillin LD motifs
by the Focal Adhesion Targeting Domain
Structure, 11, 2003
|
|
2VF2
 
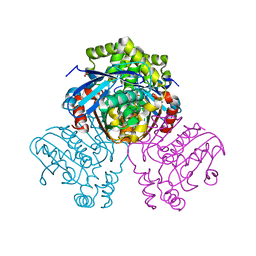 | | X-ray crystal structure of HsaD from Mycobacterium tuberculosis | | Descriptor: | 2-HYDROXY-6-OXO-6-PHENYLHEXA-2,4-DIENOATE HYDROLASE BPHD, GLYCEROL, SULFATE ION | | Authors: | Lack, N, Lowe, E.D, Liu, J, Eltis, L.D, Noble, M.E.M, Sim, E, Westwood, I.M. | | Deposit date: | 2007-10-29 | | Release date: | 2007-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of Hsad, a Steroid-Degrading Hydrolase, from Mycobacterium Tuberculosis.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
2V9C
 
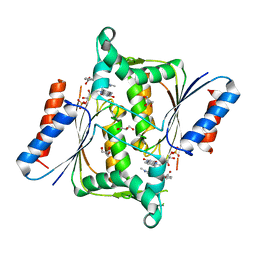 | | X-ray Crystallographic Structure of a Pseudomonas aeruginosa Azoreductase in Complex with Methyl Red. | | Descriptor: | 2-(4-DIMETHYLAMINOPHENYL)DIAZENYLBENZOIC ACID, FLAVIN MONONUCLEOTIDE, FMN-DEPENDENT NADH-AZOREDUCTASE 1, ... | | Authors: | Wang, C.-J, Hagemeier, C, Rahman, N, Lowe, E.D, Noble, M.E.M, Coughtrie, M, Sim, E, Westwood, I.M. | | Deposit date: | 2007-08-23 | | Release date: | 2007-11-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Molecular Cloning, Characterisation and Ligand- Bound Structure of an Azoreductase from Pseudomonas Aeruginosa
J.Mol.Biol., 373, 2007
|
|
2VFC
 
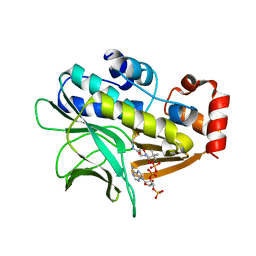 | | The structure of Mycobacterium marinum arylamine N-acetyltransferase in complex with CoA | | Descriptor: | ARYLAMINE N-ACETYLTRANSFERASE, COENZYME A | | Authors: | Fullam, E, Westwood, I.M, Anderton, M.C, Lowe, E.D, Sim, E, Noble, M.E.M. | | Deposit date: | 2007-11-02 | | Release date: | 2007-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Divergence of Cofactor Recognition Across Evolution: Coenzyme a Binding in a Prokaryotic Arylamine N-Acetyltransferase.
J.Mol.Biol., 375, 2008
|
|
2VZC
 
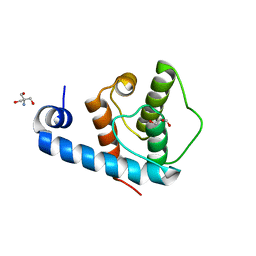 | | Crystal structure of the C-terminal calponin homology domain of alpha parvin | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Lorenz, S, Vakonakis, I, Lowe, E.D, Campbell, I.D, Noble, M.E.M, Hoellerer, M.K. | | Deposit date: | 2008-07-31 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural Analysis of the Interactions between Paxillin Ld Motifs and Alpha-Parvin
Structure, 16, 2008
|
|
2VZD
 
 | | Crystal structure of the C-terminal calponin homology domain of alpha parvin in complex with paxillin LD1 motif | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-PARVIN, GLYCEROL, ... | | Authors: | Lorenz, S, Vakonakis, I, Lowe, E.D, Campbell, I.D, Noble, M.E.M, Hoellerer, M.K. | | Deposit date: | 2008-07-31 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Analysis of the Interactions between Paxillin Ld Motifs and Alpha-Parvin
Structure, 16, 2008
|
|
