2F42
 
 | |
8SUV
 
 | | CHIP-TPR in complex with the C-terminus of CHIC2 | | 分子名称: | Cysteine-rich hydrophobic domain-containing protein 2, E3 ubiquitin-protein ligase CHIP, SULFATE ION | | 著者 | Cupo, A.R, McDermott, L.E, DeSilva, A.R, Callahan, M, Nix, J.C, Gestwicki, J.E, Page, R.C. | | 登録日 | 2023-05-13 | | 公開日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Interaction with the membrane-anchored protein CHIC2 constrains the ubiquitin ligase activity of CHIP
Biorxiv, 2023
|
|
7UOF
 
 | | Dihydroorotase from M. jannaschii | | 分子名称: | Dihydroorotase, ZINC ION | | 著者 | Vitali, J, Nix, J.C, Newman, H.E, Colaneri, M.J. | | 登録日 | 2022-04-12 | | 公開日 | 2022-08-31 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of Methanococcus jannaschii dihydroorotase.
Proteins, 91, 2023
|
|
8D3F
 
 | | Crystal structure of human STAT1 in complex with the repeat region from Toxoplasma protein TgIST | | 分子名称: | Signal transducer and activator of transcription 1-alpha/beta,Inhibitor of STAT1-dependent transcription TgIST | | 著者 | Huang, Z, Liu, H, Nix, J.C, Amarasinghe, G.K, Sibley, L.D. | | 登録日 | 2022-06-01 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.97 Å) | | 主引用文献 | The intrinsically disordered protein TgIST from Toxoplasma gondii inhibits STAT1 signaling by blocking cofactor recruitment.
Nat Commun, 13, 2022
|
|
1DDY
 
 | |
2OXQ
 
 | | Structure of the UbcH5 :CHIP U-box complex | | 分子名称: | CHLORIDE ION, STIP1 homology and U-Box containing protein 1, Ubiquitin-conjugating enzyme E2D 1 | | 著者 | Xu, Z, Nix, J.C, Devlin, K.I, Misra, S. | | 登録日 | 2007-02-20 | | 公開日 | 2008-02-19 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Interactions between the quality control ubiquitin ligase CHIP and ubiquitin conjugating enzymes.
Bmc Struct.Biol., 8, 2008
|
|
6D3P
 
 | | Crystal structure of an exoribonuclease-resistant RNA from Sweet clover necrotic mosaic virus (SCNMV) | | 分子名称: | IRIDIUM HEXAMMINE ION, RNA (45-MER) | | 著者 | Steckelberg, A.-L, Akiyama, B.M, Costantino, D.A, Sit, T.L, Nix, J.C, Kieft, J.S. | | 登録日 | 2018-04-16 | | 公開日 | 2018-06-20 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | A folded viral noncoding RNA blocks host cell exoribonucleases through a conformationally dynamic RNA structure.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6NHK
 
 | | Mortalin nucleotide binding domain in the ADP-bound state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Page, R.C, Moseng, M.A, Nix, J.C. | | 登録日 | 2018-12-23 | | 公開日 | 2019-04-17 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.777 Å) | | 主引用文献 | Biophysical Consequences of EVEN-PLUS Syndrome Mutations for the Function of Mortalin.
J.Phys.Chem.B, 123, 2019
|
|
7M7W
 
 | | Antibodies to the SARS-CoV-2 receptor-binding domain that maximize breadth and resistance to viral escape | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Monoclonal antibody S2H97 Fab heavy chain, Monoclonal antibody S2H97 Fab light chain, ... | | 著者 | Snell, G, Czudnochowski, N, Croll, T.I, Nix, J.C, Corti, D, Cameroni, E, Pinto, D, Beltramello, M. | | 登録日 | 2021-03-29 | | 公開日 | 2021-05-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape.
Nature, 597, 2021
|
|
7M98
 
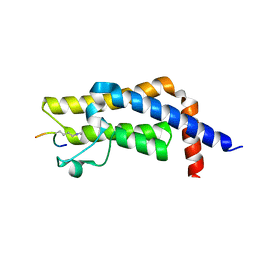 | | ATAD2 bromodomain complexed with histone H4K5ac (res 1-10) ligand | | 分子名称: | ATPase family AAA domain-containing protein 2, Histone H4 | | 著者 | Malone, K.L, Phillips, M, Nix, J.C, Glass, K.C. | | 登録日 | 2021-03-30 | | 公開日 | 2021-09-22 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Coordination of Di-Acetylated Histone Ligands by the ATAD2 Bromodomain.
Int J Mol Sci, 22, 2021
|
|
2QVU
 
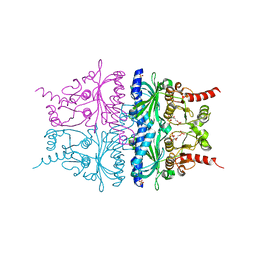 | | Porcine Liver Fructose-1,6-bisphosphatase cocrystallized with Fru-2,6-P2 and Mg2+, I(T)-state | | 分子名称: | 2,6-di-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase 1, MAGNESIUM ION, ... | | 著者 | Hines, J.K, Chen, X, Nix, J.C, Fromm, H.J, Honzatko, R.B. | | 登録日 | 2007-08-08 | | 公開日 | 2007-10-23 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structures of mammalian and bacterial fructose-1,6-bisphosphatase reveal the basis for synergism in AMP/fructose 2,6-bisphosphate inhibition
J.Biol.Chem., 282, 2007
|
|
2QVV
 
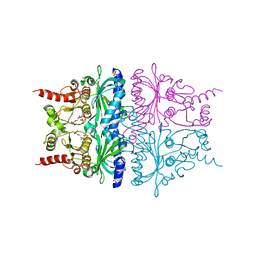 | | Porcine Liver Fructose-1,6-bisphosphatase cocrystallized with Fru-2,6-P2 and Zn2+, I(T)-state | | 分子名称: | 2,6-di-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase 1, PHOSPHATE ION, ... | | 著者 | Hines, J.K, Chen, X, Nix, J.C, Fromm, H.J, Honzatko, R.B. | | 登録日 | 2007-08-08 | | 公開日 | 2007-10-23 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Structures of mammalian and bacterial fructose-1,6-bisphosphatase reveal the basis for synergism in AMP/fructose 2,6-bisphosphate inhibition
J.Biol.Chem., 282, 2007
|
|
7R6X
 
 | | SARS-CoV-2 spike receptor-binding domain (RBD) in complex with S2E12 Fab, S309 Fab, and S304 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Monoclonal antibody S2E12 Fab heavy chain, ... | | 著者 | Snell, G, Czudnochowski, N, Croll, T.I, Nix, J.C, Corti, D, Cameroni, E, Pinto, D, Beltramello, M. | | 登録日 | 2021-06-23 | | 公開日 | 2021-07-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape.
Nature, 597, 2021
|
|
6MJ0
 
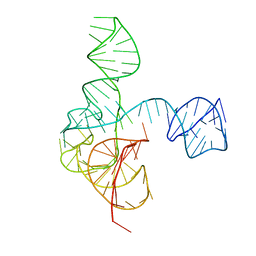 | | Crystal structure of the complete turnip yellow mosaic virus 3'UTR | | 分子名称: | RNA (101-MER) | | 著者 | Hartwick, E.W, Costantino, D.A, MacFadden, A, Nix, J.C, Tian, S, Das, R, Kieft, J.S. | | 登録日 | 2018-09-20 | | 公開日 | 2019-01-09 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Ribosome-induced RNA conformational changes in a viral 3'-UTR sense and regulate translation levels.
Nat Commun, 9, 2018
|
|
7R6W
 
 | | SARS-CoV-2 spike receptor-binding domain (RBD) in complex with S2X35 Fab and S309 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | 著者 | Snell, G, Czudnochowski, N, Hernandez, P, Nix, J.C, Croll, T.I, Corti, D, Cameroni, E, Pinto, D, Beltramello, M. | | 登録日 | 2021-06-23 | | 公開日 | 2021-07-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape.
Nature, 597, 2021
|
|
6OP6
 
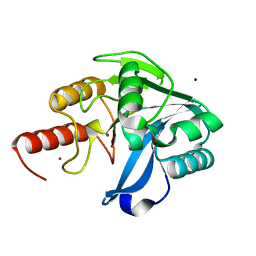 | | Structure of VIM-20 in the reduced state | | 分子名称: | Metallo-beta-lactamase VIM-20, SODIUM ION, ZINC ION | | 著者 | Page, R.C, Shurina, B.A, Montgomery, J.S, Orischak, M.G, Nix, J.C. | | 登録日 | 2019-04-24 | | 公開日 | 2019-10-23 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | A Single Salt Bridge in VIM-20 Increases Protein Stability and Antibiotic Resistance under Low-Zinc Conditions.
Mbio, 10, 2019
|
|
5GYJ
 
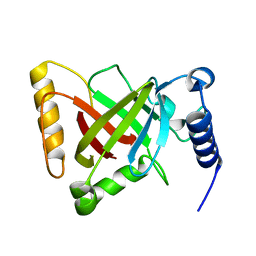 | | Structure of catalytically active sortase from Clostridium difficile | | 分子名称: | Putative peptidase C60B, sortase B | | 著者 | Yin, J.-C, Fei, C.-H, Hsiao, Y.-Y, Nix, J.C, Huang, I.-H, Wang, S. | | 登録日 | 2016-09-22 | | 公開日 | 2017-01-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.801 Å) | | 主引用文献 | Structural Insights into Substrate Recognition by Clostridium difficile Sortase.
Front Cell Infect Microbiol, 6, 2016
|
|
6OP7
 
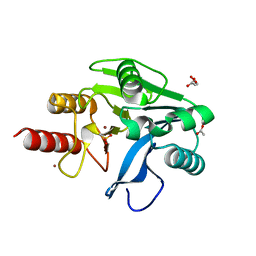 | | Structure of oxidized VIM-20 | | 分子名称: | ACETATE ION, Metallo-beta-lactamase VIM-20, ZINC ION | | 著者 | Page, R.C, Shurina, B.A, Montgomery, J.S, Orischak, M.G, Nix, J.C. | | 登録日 | 2019-04-24 | | 公開日 | 2019-10-23 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.37 Å) | | 主引用文献 | A Single Salt Bridge in VIM-20 Increases Protein Stability and Antibiotic Resistance under Low-Zinc Conditions.
Mbio, 10, 2019
|
|
6PMT
 
 | |
7TN0
 
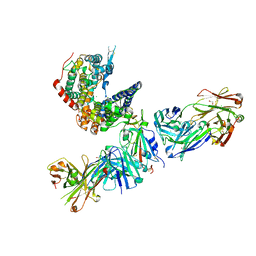 | | SARS-CoV-2 Omicron RBD in complex with human ACE2 and S304 Fab and S309 Fab | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | McCallum, M, Czudnochowski, N, Nix, J.C, Croll, T.I, SSGCID, Dillen, J.R, Snell, G, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2022-01-20 | | 公開日 | 2022-02-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Structural basis of SARS-CoV-2 Omicron immune evasion and receptor engagement.
Science, 375, 2022
|
|
7L0N
 
 | | Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity | | 分子名称: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Snell, G, Czudnochowski, N, Dillen, J, Nix, J.C, Croll, T.I, Corti, D. | | 登録日 | 2020-12-11 | | 公開日 | 2021-02-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.78 Å) | | 主引用文献 | Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity.
Cell, 184, 2021
|
|
7JJU
 
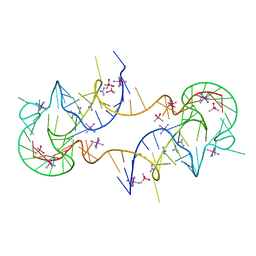 | | Crystal structure of en exoribonuclease-resistant RNA (xrRNA) from Potato leafroll virus (PLRV) | | 分子名称: | CACODYLATE ION, Guanidinium, IRIDIUM HEXAMMINE ION, ... | | 著者 | Steckelberg, A.-L, Vicens, Q, Auffinger, P, Costantino, D.C, Nix, J.C, Kieft, J.S. | | 登録日 | 2020-07-27 | | 公開日 | 2020-09-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.604 Å) | | 主引用文献 | The crystal structure of a Polerovirus exoribonuclease-resistant RNA shows how diverse sequences are integrated into a conserved fold.
Rna, 26, 2020
|
|
7JX3
 
 | | Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab domain of monoclonal antibody S2H14, Heavy chain of Fab domain of monoclonal antibody S304, ... | | 著者 | Snell, G, Czudnochowski, N, Rosen, L.E, Nix, J.C, Corti, D, Veesler, D, Park, Y.J, Walls, A.C, Tortorici, M.A, Cameroni, E, Pinto, D, Beltramello, M, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2020-08-26 | | 公開日 | 2020-10-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
5TPY
 
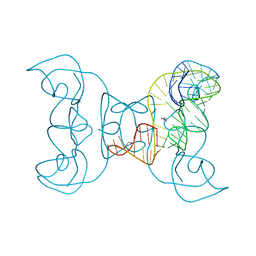 | | Crystal structure of an exonuclease resistant RNA from Zika virus | | 分子名称: | HEXANE-1,6-DIOL, MAGNESIUM ION, RNA (71-MER) | | 著者 | Akiyama, B.M, Laurence, H.M, Massey, A.R, Costantino, D.A, Xie, X, Yang, Y, Shi, P.-Y, Nix, J.C, Beckham, J.D, Kieft, J.S. | | 登録日 | 2016-10-21 | | 公開日 | 2016-12-14 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.805 Å) | | 主引用文献 | Zika virus produces noncoding RNAs using a multi-pseudoknot structure that confounds a cellular exonuclease.
Science, 354, 2016
|
|
5F14
 
 | |
