6LKT
 
 | | Crystal structure of the Fab fragment of murine monoclonal antibody KH-1 against Human herpesvirus 6B | | Descriptor: | antibody Fab Fragment L-chain, antibody Fab fragment H chain | | Authors: | Nishimura, M, Novita, B.D, Kato, T, Tjan, L.H, Wang, B, Wakata, A, Poetranto, A.L, Kawabata, A, Tang, H, Aoshi, T, Mori, Y. | | Deposit date: | 2019-12-20 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the interaction of human herpesvirus 6B tetrameric glycoprotein complex with the cellular receptor, human CD134.
Plos Pathog., 16, 2020
|
|
3C3I
 
 | | Evolution of chlorella virus dUTPase | | Descriptor: | DEOXYURIDINE-5'-DIPHOSPHATE, Deoxyuridine triphosphatase, MAGNESIUM ION | | Authors: | Yamanishi, M, Homma, K, Zhang, Y, Etten, L.V.J, Moriyama, H. | | Deposit date: | 2008-01-28 | | Release date: | 2009-02-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallization and crystal-packing studies of Chlorella virus deoxyuridine triphosphatase.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
1WKI
 
 | | solution structure of ribosomal protein L16 from thermus thermophilus HB8 | | Descriptor: | LSU ribosomal protein L16P | | Authors: | Nishimura, M, Yoshida, T, Shirouzu, M, Terada, T, Kuramitsu, S, Yokoyama, S, Ohkubo, T, Kobayashi, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-31 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Ribosomal Protein L16 from Thermus thermophilus HB8
J.Mol.Biol., 344, 2004
|
|
3CA9
 
 | | Evolution of chlorella virus dUTPase | | Descriptor: | DEOXYURIDINE-5'-DIPHOSPHATE, Deoxyuridine triphosphatase, MAGNESIUM ION | | Authors: | Yamanishi, M, Homma, K, Zhang, Y, Etten, L.V.J, Moriyama, H. | | Deposit date: | 2008-02-19 | | Release date: | 2009-03-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallization and crystal-packing studies of Chlorella virus deoxyuridine triphosphatase.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3C2T
 
 | | Evolution of chlorella virus dUTPase | | Descriptor: | DEOXYURIDINE-5'-DIPHOSPHATE, Deoxyuridine triphosphatase, MAGNESIUM ION | | Authors: | Yamanishi, M, Homma, K, Zhang, Y, Etten, L.V.J, Moriyama, H. | | Deposit date: | 2008-01-25 | | Release date: | 2009-02-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallization and crystal-packing studies of Chlorella virus deoxyuridine triphosphatase.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
1X23
 
 | | Crystal structure of ubch5c | | Descriptor: | Ubiquitin-conjugating enzyme E2 D3 | | Authors: | Nakanishi, M, Teshima, N, Mizushima, T, Murata, S, Tanaka, K, Yamane, T. | | Deposit date: | 2005-04-19 | | Release date: | 2005-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of ubch5c
To be Published
|
|
1Q2H
 
 | | Phenylalanine Zipper Mediates APS Dimerization | | Descriptor: | adaptor protein with pleckstrin homology and src homology 2 domains | | Authors: | Dhe-Paganon, S, Werner, E.D, Nishi, M, Chi, Y.-I, Shoelson, S.E. | | Deposit date: | 2003-07-24 | | Release date: | 2004-08-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A phenylalanine zipper mediates APS dimerization.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1JQE
 
 | | Crystal Structure Analysis of Human Histamine Methyltransferase (Ile105 Polymorphic Variant) Complexed with AdoHcy and Antimalarial Drug Quinacrine | | Descriptor: | Histamine N-Methyltransferase, QUINACRINE, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Horton, J.R, Sawada, K, Nishibori, M, Zhang, X, Cheng, X. | | Deposit date: | 2001-08-06 | | Release date: | 2002-08-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Two polymorphic forms of human histamine methyltransferase: structural, thermal, and kinetic comparisons.
Structure, 9, 2001
|
|
8IP8
 
 | | Wheat 80S ribosome stalled on AUG-Stop boron dependently | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein eL8, 40S ribosomal protein eS1, ... | | Authors: | Yokoyama, T, Tanaka, M, Saito, H, Nishimoto, M, Tsuda, K, Sotta, N, Shigematsu, H, Shirouzu, M, Iwasaki, S, Ito, T, Fujiwara, T. | | Deposit date: | 2023-03-14 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Boric acid intercepts 80S ribosome migration from AUG-stop by stabilizing eRF1.
Nat.Chem.Biol., 20, 2024
|
|
1K1Z
 
 | | Solution structure of N-terminal SH3 domain mutant(P33G) of murine Vav | | Descriptor: | vav | | Authors: | Ogura, K, Nagata, K, Horiuchi, M, Ebisui, E, Hasuda, T, Yuzawa, S, Nishida, M, Hatanaka, H, Inagaki, F. | | Deposit date: | 2001-09-26 | | Release date: | 2001-10-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of N-terminal SH3 domain of Vav and the recognition site for Grb2 C-terminal SH3 domain
J.BIOMOL.NMR, 22, 2002
|
|
6IPW
 
 | | Crystal structure of CqsB2 from Streptomyces exfoliatus in complex with the product, 1-(2-hydroxypropyl)-2-methyl-carbazole-3,4-dione | | Descriptor: | 2-methyl-1-[(2R)-2-oxidanylpropyl]-9H-carbazole-3,4-dione, CqsB2 | | Authors: | Tomita, T, Kobayashi, M, Nishiyama, M, Kuzuyama, T. | | Deposit date: | 2018-11-05 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An Unprecedented Cyclization Mechanism in the Biosynthesis of Carbazole Alkaloids in Streptomyces.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6IPV
 
 | |
6ITD
 
 | | Crystal structure of BioU (K124A) from Synechocystis sp.PCC6803 in complex with the analog of reaction intermediate, 3-(1-aminoethyl)-nonanedioic acid | | Descriptor: | 3-(1-AMINOETHYL)NONANEDIOIC ACID, Slr0355 protein | | Authors: | Sakaki, K, Tomita, T, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2018-11-21 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A suicide enzyme catalyzes multiple reactions for biotin biosynthesis in cyanobacteria.
Nat.Chem.Biol., 16, 2020
|
|
8J3T
 
 | | Complex structure of human cytomegalovirus protease and a non-covalent small-molecule ligand | | Descriptor: | (4R)-1-[1-[(S)-[1-cyclopentyl-3-(2-methylphenyl)pyrazol-4-yl]-(4-methylphenyl)methyl]-2-oxidanylidene-pyridin-3-yl]-3-methyl-2-oxidanylidene-N-(3-oxidanylidene-2-azabicyclo[2.2.2]octan-4-yl)imidazolidine-4-carboxamide, Assemblin | | Authors: | Yoshida, S, Sako, Y, Nikaido, E, Ueda, T, Kozono, I, Ichihashi, Y, Nakahashi, A, Onishi, M, Yamatsu, Y, Kato, T, Nishikawa, J, Tachibana, Y. | | Deposit date: | 2023-04-18 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Peptide-to-Small Molecule: Discovery of Non-Covalent, Active-Site Inhibitors of beta-Herpesvirus Proteases.
Acs Med.Chem.Lett., 14, 2023
|
|
8J3S
 
 | | Complex structure of human cytomegalovirus protease and a macrocyclic peptide ligand | | Descriptor: | Assemblin, PHE-ILE-THR-GLY-HIS-TYR-TRP-VAL-ARG-PHE-LEU-PRO-CYS-GLY | | Authors: | Yoshida, S, Sako, Y, Nikaido, E, Ueda, T, Kozono, I, Ichihashi, Y, Nakahashi, A, Onishi, M, Yamatsu, Y, Kato, T, Nishikawa, J, Tachibana, Y. | | Deposit date: | 2023-04-18 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Peptide-to-Small Molecule: Discovery of Non-Covalent, Active-Site Inhibitors of beta-Herpesvirus Proteases.
Acs Med.Chem.Lett., 14, 2023
|
|
5WVO
 
 | | Crystal structure of DNMT1 RFTS domain in complex with K18/K23 mono-ubiquitylated histone H3 | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, Histone H3.1, Ubiquitin, ... | | Authors: | Ishiyama, S, Nishiyama, A, Nakanishi, M, Arita, K. | | Deposit date: | 2016-12-28 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structure of the Dnmt1 Reader Module Complexed with a Unique Two-Mono-Ubiquitin Mark on Histone H3 Reveals the Basis for DNA Methylation Maintenance
Mol. Cell, 68, 2017
|
|
8XZ2
 
 | | The structural model of a homodimeric D-Ala-D-Ala metallopeptidase, VanX, from vancomycin-resistant bacteria | | Descriptor: | D-alanyl-D-alanine dipeptidase | | Authors: | Konuma, T, Takai, T, Tsuchiya, C, Nishida, M, Hashiba, M, Yamada, Y, Shirai, H, Motoda, Y, Nagadoi, A, Chikaishi, E, Akagi, K, Akashi, S, Yamazaki, T, Akutsu, H, Oe, A, Ikegami, T. | | Deposit date: | 2024-01-20 | | Release date: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Analysis of the homodimeric structure of a D-Ala-D-Ala metallopeptidase, VanX, from vancomycin-resistant bacteria.
Protein Sci., 33, 2024
|
|
8IP9
 
 | | Wheat 40S ribosome in complex with a tRNAi | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S23, 40S ribosomal protein eS1, ... | | Authors: | Yokoyama, T, Tanaka, M, Saito, H, Nishimoto, M, Tsuda, K, Sotta, N, Shigematsu, H, Shirouzu, M, Iwasaki, S, Ito, T, Fujiwara, T. | | Deposit date: | 2023-03-14 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Boric acid intercepts 80S ribosome migration from AUG-stop by stabilizing eRF1.
Nat.Chem.Biol., 20, 2024
|
|
8IPB
 
 | | Wheat 80S ribosome pausing on AUG-Stop with cycloheximide | | Descriptor: | 18S ribosomal RNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, 40S ribosomal protein eL8, ... | | Authors: | Yokoyama, T, Tanaka, M, Saito, H, Nishimoto, M, Tsuda, K, Sotta, N, Shigematsu, H, Shirouzu, M, Iwasaki, S, Ito, T, Fujiwara, T. | | Deposit date: | 2023-03-14 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Boric acid intercepts 80S ribosome migration from AUG-stop by stabilizing eRF1.
Nat.Chem.Biol., 20, 2024
|
|
8IPA
 
 | | Wheat 80S ribosome stalled on AUG-Stop boron dependently with cycloheximide | | Descriptor: | 18S ribosomal RNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, 40S ribosomal protein eL8, ... | | Authors: | Yokoyama, T, Tanaka, M, Saito, H, Nishimoto, M, Tsuda, K, Sotta, N, Shigematsu, H, Shirouzu, M, Iwasaki, S, Ito, T, Fujiwara, T. | | Deposit date: | 2023-03-14 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Boric acid intercepts 80S ribosome migration from AUG-stop by stabilizing eRF1.
Nat.Chem.Biol., 20, 2024
|
|
8XI6
 
 | | SARS-CoV-2 Omicron BQ.1.1 Variant Spike Protein Complexed with MO11 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ishimaru, H, Nishimura, M, Shigematsu, H, Marini, M.I, Hasegawa, N, Takamiya, R, Iwata, S, Mori, Y. | | Deposit date: | 2023-12-19 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Epitopes of an antibody that neutralizes a wide range of SARS-CoV-2 variants in a conserved subdomain 1 of the spike protein.
J.Virol., 98, 2024
|
|
3A3U
 
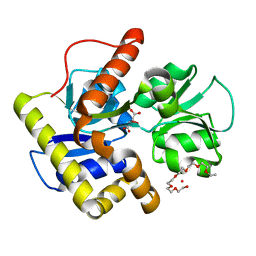 | | Crystal structure of MqnD (TTHA1568), a menaquinone biosynthetic enzyme from Thermus thermophilus HB8 | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, L(+)-TARTARIC ACID, Menaquinone biosynthetic enzyme, ... | | Authors: | Arai, R, Nishino, A, Nagano, K, Uchikubo-Kamo, T, Katsura, K, Nishimoto, M, Toyama, M, Terada, T, Kuramitsu, S, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-06-20 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of MqnD (TTHA1568), a menaquinone biosynthetic enzyme from Thermus thermophilus HB8.
J.Struct.Biol., 168, 2009
|
|
4WH2
 
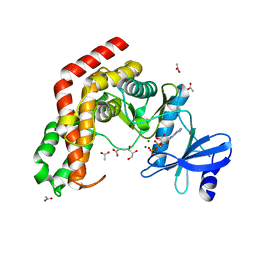 | | N-acetylhexosamine 1-kinase in complex with ADP | | Descriptor: | ACETIC ACID, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Sato, M, Arakawa, T, Nam, Y.W, Nishimoto, M, Kitaoka, M, Fushinobu, S. | | Deposit date: | 2014-09-19 | | Release date: | 2015-02-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Open-close structural change upon ligand binding and two magnesium ions required for the catalysis of N-acetylhexosamine 1-kinase
Biochim.Biophys.Acta, 1854, 2015
|
|
4WH1
 
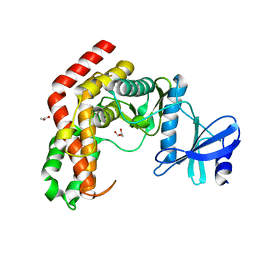 | | N-Acetylhexosamine 1-kinase (ligand free) | | Descriptor: | ACETIC ACID, GLYCEROL, N-acetylhexosamine 1-kinase | | Authors: | Sato, M, Arakawa, T, Nam, Y.W, Nishimoto, M, Kitaoka, M, Fushinobu, S. | | Deposit date: | 2014-09-19 | | Release date: | 2015-02-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Open-close structural change upon ligand binding and two magnesium ions required for the catalysis of N-acetylhexosamine 1-kinase
Biochim.Biophys.Acta, 1854, 2015
|
|
4WH3
 
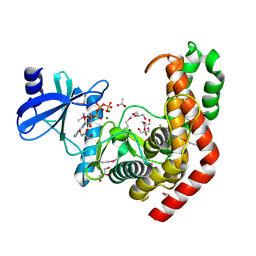 | | N-acetylhexosamine 1-kinase in complex with ATP | | Descriptor: | ACETIC ACID, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Sato, M, Arakawa, T, Nam, Y.W, Nishimoto, M, Kitaoka, M, Fushinobu, S. | | Deposit date: | 2014-09-19 | | Release date: | 2015-02-18 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Open-close structural change upon ligand binding and two magnesium ions required for the catalysis of N-acetylhexosamine 1-kinase
Biochim.Biophys.Acta, 1854, 2015
|
|
