2Y32
 
 | | Crystal structure of bradavidin | | Descriptor: | BLR5658 PROTEIN | | Authors: | Leppiniemi, J, Gronroos, T, Johnson, M.S, Kulomaa, M.S, Hytonen, V.P, Airenne, T.T. | | Deposit date: | 2010-12-17 | | Release date: | 2011-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of Bradavidin - C-Terminal Residues Act as Intrinsic Ligands.
Plos One, 7, 2012
|
|
3KG0
 
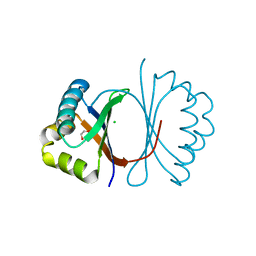 | | Crystal structure of SnoaB, a cofactor-independent oxygenase from Streptomyces nogalater, determined to 1.7 resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SnoaB | | Authors: | Koskiniemi, H, Grocholski, T, Lindqvist, Y, Mantsala, P, Niemi, J, Schneider, G. | | Deposit date: | 2009-10-28 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the cofactor-independent monooxygenase SnoaB from Streptomyces nogalater: implications for the reaction mechanism
Biochemistry, 49, 2010
|
|
3KG1
 
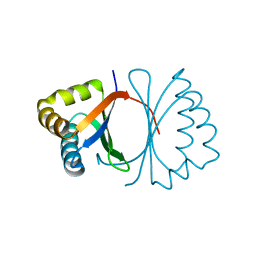 | | Crystal structure of SnoaB, a cofactor-independent oxygenase from Streptomyces nogalater, mutant N63A | | Descriptor: | CHLORIDE ION, SnoaB | | Authors: | Koskiniemi, H, Grocholski, T, Lindqvist, Y, Mantsala, P, Niemi, J, Schneider, G. | | Deposit date: | 2009-10-28 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the cofactor-independent monooxygenase SnoaB from Streptomyces nogalater: implications for the reaction mechanism
Biochemistry, 49, 2010
|
|
2QA1
 
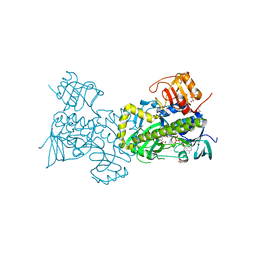 | | Crystal structure of PgaE, an aromatic hydroxylase involved in angucycline biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Koskiniemi, H, Dobritzsch, D, Metsa-Ketela, M, Kallio, P, Niemi, J, Schneider, G. | | Deposit date: | 2007-06-14 | | Release date: | 2007-08-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of two aromatic hydroxylases involved in the early tailoring steps of angucycline biosynthesis
J.Mol.Biol., 372, 2007
|
|
2QA2
 
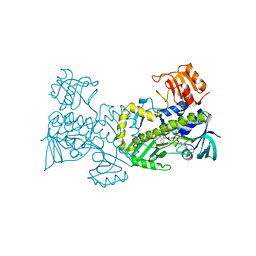 | | Crystal structure of CabE, an aromatic hydroxylase from angucycline biosynthesis, determined to 2.7 A resolution | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Polyketide oxygenase CabE | | Authors: | Koskiniemi, H, Dobritzsch, D, Metsa-Ketela, M, Kallio, P, Niemi, J, Schneider, G. | | Deposit date: | 2007-06-14 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of two aromatic hydroxylases involved in the early tailoring steps of angucycline biosynthesis
J.Mol.Biol., 372, 2007
|
|
1QZZ
 
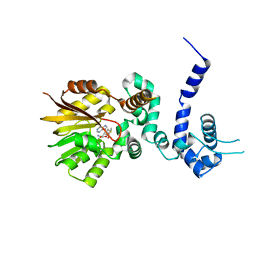 | | Crystal structure of aclacinomycin-10-hydroxylase (RdmB) in complex with S-adenosyl-L-methionine (SAM) | | Descriptor: | ACETATE ION, S-ADENOSYLMETHIONINE, aclacinomycin-10-hydroxylase | | Authors: | Jansson, A, Niemi, J, Lindqvist, Y, Mantsala, P, Schneider, G, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-09-19 | | Release date: | 2003-11-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Aclacinomycin-10-Hydroxylase, a S-Adenosyl-L-Methionine-dependent Methyltransferase Homolog Involved in Anthracycline Biosynthesis in Streptomyces purpurascens.
J.Mol.Biol., 334, 2003
|
|
1R00
 
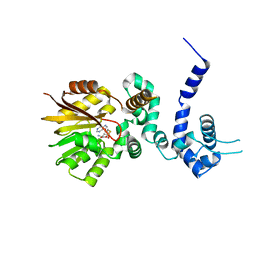 | | Crystal structure of aclacinomycin-10-hydroxylase (RdmB) in complex with S-adenosyl-L-homocysteine (SAH) | | Descriptor: | ACETATE ION, S-ADENOSYL-L-HOMOCYSTEINE, aclacinomycin-10-hydroxylase | | Authors: | Jansson, A, Niemi, J, Lindqvist, Y, Mantsala, P, Schneider, G. | | Deposit date: | 2003-09-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Aclacinomycin-10-Hydroxylase, a S-Adenosyl-L-Methionine-dependent Methyltransferase Homolog Involved in Anthracycline Biosynthesis in Streptomyces purpurascens.
J.Mol.Biol., 334, 2003
|
|
3KNG
 
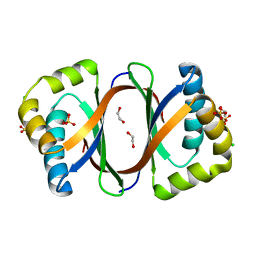 | | Crystal structure of SnoaB, a cofactor-independent oxygenase from Streptomyces nogalater, determined to 1.9 resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Koskiniemi, H, Grocholski, T, Lindqvist, Y, Mantsala, P, Niemi, J, Schneider, G. | | Deposit date: | 2009-11-12 | | Release date: | 2010-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the cofactor-independent monooxygenase SnoaB from Streptomyces nogalater: implications for the reaction mechanism
Biochemistry, 49, 2010
|
|
1XDS
 
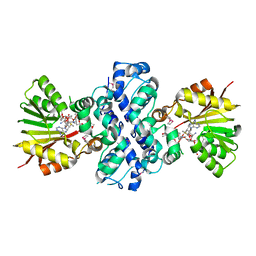 | | Crystal structure of Aclacinomycin-10-hydroxylase (RdmB) in complex with S-adenosyl-L-methionine (SAM) and 11-deoxy-beta-rhodomycin (DbrA) | | Descriptor: | 11-DEOXY-BETA-RHODOMYCIN, Protein RdmB, S-ADENOSYLMETHIONINE | | Authors: | Jansson, A, Koskiniemi, H, Erola, A, Wang, J, Mantsala, P, Schneider, G, Niemi, J, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-09-08 | | Release date: | 2004-11-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Aclacinomycin 10-Hydroxylase Is a Novel Substrate-assisted Hydroxylase Requiring S-Adenosyl-L-methionine as Cofactor
J.Biol.Chem., 280, 2005
|
|
1XDU
 
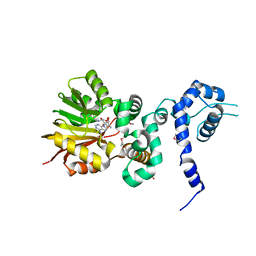 | | Crystal structure of Aclacinomycin-10-hydroxylase (RdmB) in complex with Sinefungin (SFG) | | Descriptor: | ACETATE ION, Protein RdmB, SINEFUNGIN | | Authors: | Jansson, A, Koskiniemi, H, Erola, A, Wang, J, Mantsala, P, Schneider, G, Niemi, J. | | Deposit date: | 2004-09-08 | | Release date: | 2004-11-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Aclacinomycin 10-Hydroxylase Is a Novel Substrate-assisted Hydroxylase Requiring S-Adenosyl-L-methionine as Cofactor
J.Biol.Chem., 280, 2005
|
|
1TW2
 
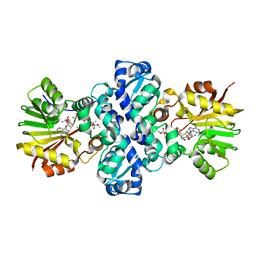 | | Crystal structure of Carminomycin-4-O-methyltransferase (DnrK) in complex with S-adenosyl-L-homocystein (SAH) and 4-methoxy-e-rhodomycin T (M-ET) | | Descriptor: | Carminomycin 4-O-methyltransferase, METHYL (4R)-2-ETHYL-2,5,12-TRIHYDROXY-7-METHOXY-6,11-DIOXO-4-{[2,3,6-TRIDEOXY-3-(DIMETHYLAMINO)-BETA-D-RIBO-HEXOPYRANOSYL]OXY}-1H,2H,3H,4H,6H,11H-TETRACENE-1-CARBOXYLATE, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Jansson, A, Koskiniemi, H, Mantsala, P, Niemi, J, Schneider, G, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-06-30 | | Release date: | 2004-09-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a ternary complex of DnrK, a methyltransferase in daunorubicin biosynthesis, with bound products
J.Biol.Chem., 279, 2004
|
|
1TW3
 
 | | Crystal structure of Carminomycin-4-O-methyltransferase (DnrK) in complex with S-adenosyl-L-homocystein (SAH) and 4-methoxy-e-rhodomycin T (M-ET) | | Descriptor: | Carminomycin 4-O-methyltransferase, METHYL (4R)-2-ETHYL-2,5,12-TRIHYDROXY-7-METHOXY-6,11-DIOXO-4-{[2,3,6-TRIDEOXY-3-(DIMETHYLAMINO)-BETA-D-RIBO-HEXOPYRANOSYL]OXY}-1H,2H,3H,4H,6H,11H-TETRACENE-1-CARBOXYLATE, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Jansson, A, Koskiniemi, H, Mantsala, P, Niemi, J, Schneider, G. | | Deposit date: | 2004-06-30 | | Release date: | 2004-09-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of a ternary complex of DnrK, a methyltransferase in daunorubicin biosynthesis, with bound products
J.Biol.Chem., 279, 2004
|
|
1Q0R
 
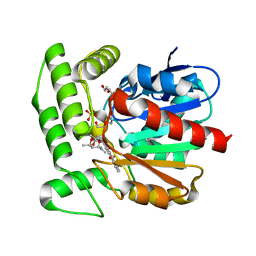 | | Crystal structure of aclacinomycin methylesterase (RdmC) with bound product analogue, 10-decarboxymethylaclacinomycin T (DcmaT) | | Descriptor: | 10-DECARBOXYMETHYLACLACINOMYCIN T (DCMAT), PENTAETHYLENE GLYCOL, SULFATE ION, ... | | Authors: | Jansson, A, Niemi, J, Mantsala, P, Schneider, G, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-07-17 | | Release date: | 2003-11-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of aclacinomycin methylesterase with bound product analogues: implications for anthracycline recognition and mechanism.
J.Biol.Chem., 278, 2003
|
|
1Q0Z
 
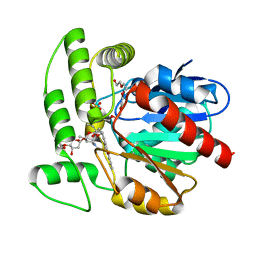 | | Crystal structure of aclacinomycin methylesterase (RdmC) with bound product analogue, 10-decarboxymethylaclacinomycin A (DcmA) | | Descriptor: | 10-DECARBOXYMETHYLACLACINOMYCIN A (DCMAA), PENTAETHYLENE GLYCOL, SULFATE ION, ... | | Authors: | Jansson, A, Niemi, J, Mantsala, P, Schneider, G, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-07-18 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of aclacinomycin methylesterase with bound product analogues: implications for anthracycline recognition and mechanism.
J.Biol.Chem., 278, 2003
|
|
2IPI
 
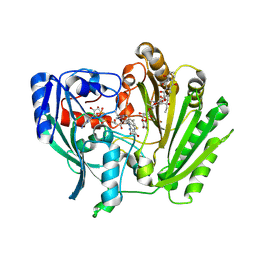 | | Crystal Structure of Aclacinomycin Oxidoreductase | | Descriptor: | Aclacinomycin oxidoreductase (AknOx), FLAVIN-ADENINE DINUCLEOTIDE, METHYL (2S,4R)-2-ETHYL-2,5,7-TRIHYDROXY-6,11-DIOXO-4-{[2,3,6-TRIDEOXY-4-O-{2,6-DIDEOXY-4-O-[(2S,6S)-6-METHYL-5-OXOTETRAHYDRO-2H-PYRAN-2-YL]-ALPHA-D-LYXO-HEXOPYRANOSYL}-3-(DIMETHYLAMINO)-D-RIBO-HEXOPYRANOSYL]OXY}-1,2,3,4,6,11-HEXAHYDROTETRACENE-1-CARBOXYLATE | | Authors: | Sultana, A, Kursula, I, Schneider, G, Alexeev, I, Niemi, J, Mantsala, P. | | Deposit date: | 2006-10-12 | | Release date: | 2007-01-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure determination by multiwavelength anomalous diffraction of aclacinomycin oxidoreductase: indications of multidomain pseudomerohedral twinning.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
4ICY
 
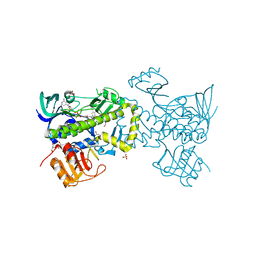 | | Tracing the Evolution of Angucyclinone Monooxygenases: Structural Determinants for C-12b Hydroxylation and Substrate Inhibition in PgaE | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kallio, P, Patrikainen, P, Belogurov, G, Mantsala, P, Yang, K, Niemi, J, Metsa-Ketela, M. | | Deposit date: | 2012-12-11 | | Release date: | 2013-06-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Tracing the Evolution of Angucyclinone Monooxygenases: Structural Determinants for C-12b Hydroxylation and Substrate Inhibition in PgaE.
Biochemistry, 52, 2013
|
|
4KWH
 
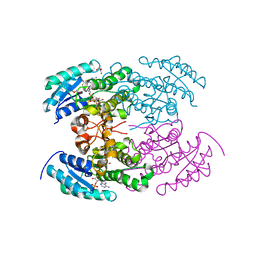 | | The crystal structure of angucycline C-6 ketoreductase LanV with bound NADP | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Paananen, P, Patrikainen, P, Kallio, P, Mantsala, P, Niemi, J, Niiranen, L, Metsa-Ketela, M. | | Deposit date: | 2013-05-24 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional analysis of angucycline C-6 ketoreductase LanV involved in landomycin biosynthesis.
Biochemistry, 52, 2013
|
|
4KWI
 
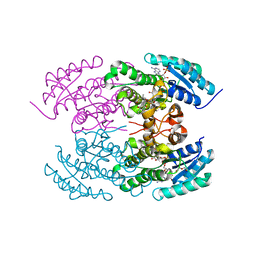 | | The crystal structure of angucycline C-6 ketoreductase LanV with bound NADP and 11-deoxy-6-oxylandomycinone | | Descriptor: | 1,8-dihydroxy-3-methyltetraphene-6,7,12(5H)-trione, DI(HYDROXYETHYL)ETHER, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Paananen, P, Patrikainen, P, Mantsala, P, Niemi, J, Niiranen, L, Metsa-Ketela, M. | | Deposit date: | 2013-05-24 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional analysis of angucycline C-6 ketoreductase LanV involved in landomycin biosynthesis.
Biochemistry, 52, 2013
|
|
4EX6
 
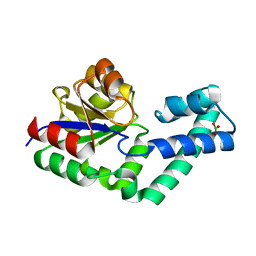 | | Crystal structure of the alnumycin P phosphatase AlnB | | Descriptor: | AlnB, BORIC ACID, MAGNESIUM ION | | Authors: | Oja, T, Niiranen, L, Sandalova, T, Klika, K.D, Niemi, J, Mantsala, P, Schneider, G, Metsa-Ketela, M. | | Deposit date: | 2012-04-30 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural basis for C-ribosylation in the alnumycin A biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4EX9
 
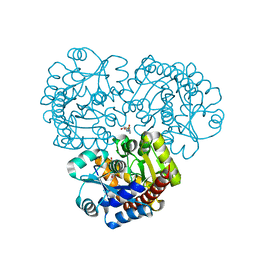 | | Crystal structure of the prealnumycin C-glycosynthase AlnA in complex with ribulose 5-phosphate | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AlnA, CALCIUM ION, ... | | Authors: | Oja, T, Niiranen, L, Sandalova, T, Klika, K.D, Niemi, J, Mantsala, P, Schneider, G, Metsa-Ketela, M. | | Deposit date: | 2012-04-30 | | Release date: | 2013-01-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis for C-ribosylation in the alnumycin A biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4EX7
 
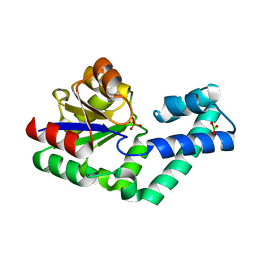 | | Crystal structure of the alnumycin P phosphatase in complex with free phosphate | | Descriptor: | AlnB, BORIC ACID, MAGNESIUM ION, ... | | Authors: | Oja, T, Niiranen, L, Sandalova, T, Klika, K.D, Niemi, J, Mantsala, P, Schneider, G, Metsa-Ketela, M. | | Deposit date: | 2012-04-30 | | Release date: | 2013-01-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for C-ribosylation in the alnumycin A biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4EX8
 
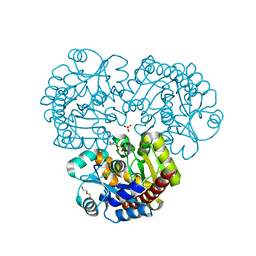 | | Crystal structure of the prealnumycin C-glycosynthase AlnA | | Descriptor: | AlnA, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Oja, T, Niiranen, L, Sandalova, T, Klika, K.D, Niemi, J, Mantsala, P, Schneider, G, Metsa-Ketela, M. | | Deposit date: | 2012-04-30 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for C-ribosylation in the alnumycin A biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
