5LCB
 
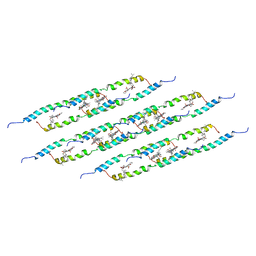 | | In situ atomic-resolution structure of the baseplate antenna complex in Chlorobaculum tepidum obtained combining solid-state NMR spectroscopy, cryo electron microscopy and polarization spectroscopy | | Descriptor: | BACTERIOCHLOROPHYLL A, Bacteriochlorophyll c-binding protein | | Authors: | Nielsen, J.T, Kulminskaya, N.V, Bjerring, M, Linnanto, J.M, Ratsep, M, Pedersen, M, Lambrev, P.H, Dorogi, M, Garab, G, Thomsen, K, Jegerschold, C, Frigaard, N.U, Lindahl, M, Nielsen, N.C. | | Deposit date: | 2016-06-20 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (26.5 Å), SOLID-STATE NMR | | Cite: | In situ high-resolution structure of the baseplate antenna complex in Chlorobaculum tepidum.
Nat Commun, 7, 2016
|
|
2QTX
 
 | | Crystal structure of an Hfq-like protein from Methanococcus jannaschii | | Descriptor: | Uncharacterized protein MJ1435 | | Authors: | Nielsen, J.S, Boggild, A, Andersen, C.B.F, Nielsen, G, Boysen, A, Brodersen, D.E, Valentin-Hansen, P. | | Deposit date: | 2007-08-03 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An Hfq-like protein in archaea: Crystal structure and functional characterization of the Sm protein from Methanococcus jannaschii.
Rna, 13, 2007
|
|
2KIB
 
 | | Protein Fibril | | Descriptor: | NFGAIL segment from human islet amyloid polypeptide | | Authors: | Nielsen, J.T, Bjerring, M, Jeppesen, M.D, Pedersen, R.O, Pedersen, J.M, Hein, K.L, Vosegaard, T, Skrydstrup, T, Otzen, D.E, Nielsen, N. | | Deposit date: | 2009-05-01 | | Release date: | 2009-09-08 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Unique identification of supramolecular structures in amyloid fibrils by solid-state NMR spectroscopy.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
9EO4
 
 | | Outward-open structure of human dopamine transporter bound to cocaine | | Descriptor: | CHLORIDE ION, CHOLESTEROL, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Nielsen, J.C, Salomon, K, Kalenderoglou, I.E, Bargmeyer, S, Pape, T, Shahsavar, A, Loland, C.J. | | Deposit date: | 2024-03-14 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structure of the human dopamine transporter in complex with cocaine.
Nature, 632, 2024
|
|
6TKT
 
 | |
7PG2
 
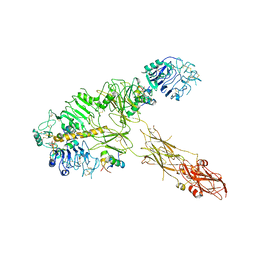 | | Low resolution Cryo-EM structure of full-length insulin receptor bound to 3 insulin, conf 1 | | Descriptor: | Insulin, Isoform Short of Insulin receptor | | Authors: | Nielsen, J.A, Slaaby, R, Boesen, T, Hummelshoj, T, Brandt, J, Schluckebier, G, Nissen, P. | | Deposit date: | 2021-08-12 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structural Investigations of Full-Length Insulin Receptor Dynamics and Signalling.
J.Mol.Biol., 434, 2022
|
|
7PG0
 
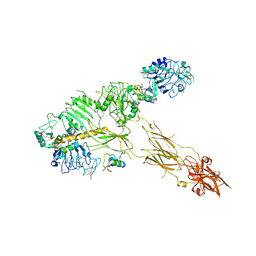 | | Low resolution Cryo-EM structure of full-length insulin receptor bound to 3 insulin with visible ddm micelle, conf 1 | | Descriptor: | Insulin, Isoform Short of Insulin receptor | | Authors: | Nielsen, J.A, Slaaby, R, Boesen, T, Hummelshoj, T, Brandt, J, Schluckebier, G, Nissen, P. | | Deposit date: | 2021-08-12 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structural Investigations of Full-Length Insulin Receptor Dynamics and Signalling.
J.Mol.Biol., 434, 2022
|
|
7PG4
 
 | | Low resolution Cryo-EM structure of the full-length insulin receptor bound to 2 insulin, conf 3 | | Descriptor: | Insulin, Isoform Short of Insulin receptor | | Authors: | Nielsen, J.A, Slaaby, R, Boesen, T, Hummelshoj, T, Brandt, J, Schluckebier, G, Nissen, P. | | Deposit date: | 2021-08-12 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | Structural Investigations of Full-Length Insulin Receptor Dynamics and Signalling.
J.Mol.Biol., 434, 2022
|
|
7PG3
 
 | | Low resolution Cryo-EM structure of the full-length insulin receptor bound to 3 insulin, conf 2 | | Descriptor: | Insulin, Isoform Short of Insulin receptor | | Authors: | Nielsen, J.A, Slaaby, R, Boesen, T, Hummelshoj, T, Brandt, J, Schluckebier, G, Nissen, P. | | Deposit date: | 2021-08-12 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Structural Investigations of Full-Length Insulin Receptor Dynamics and Signalling.
J.Mol.Biol., 434, 2022
|
|
2WCN
 
 | | Solution structure of an LNA-modified quadruplex | | Descriptor: | DNA (5'-D(*DGP*LCG*DGP*LCG*DTP*DTP*DTP *DTP*DGP*LCG*DGP*LCG)-3') | | Authors: | Nielsen, J.T, Arar, K, Petersen, M. | | Deposit date: | 2009-03-12 | | Release date: | 2009-11-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Locked Nucleic Acid Modified Quadruplex: Introducing the V4 Folding Topology.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
1OKF
 
 | | NMR structure of an alpha-L-LNA:RNA hybrid | | Descriptor: | 5'-D(*CP*ATLP*GP*AP*ATLP*AP*ATLP*GP*CP)-3', 5'-R(*GP*CP*AP*UP*AP*UP*CP*AP*GP)-3' | | Authors: | Nielsen, J.T, Stein, P.C, Petersen, M. | | Deposit date: | 2003-07-23 | | Release date: | 2003-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of an Alpha-L-Lna:RNA Hybrid: Structural Implications for Rnase H Recognition
Nucleic Acids Res., 31, 2003
|
|
2CHK
 
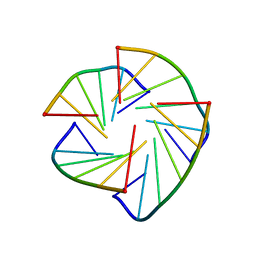 | | NMR structure of TLLLLT quadruplex | | Descriptor: | 5'-D(*T LCG LCG LCG LCGP*TP)-3' | | Authors: | Nielsen, J.T, Petersen, M. | | Deposit date: | 2006-03-15 | | Release date: | 2006-04-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structures of Lna (Locked Nucleic Acid) Modified Quadruplexes
Nucleic Acids Res., 34, 2006
|
|
2CHJ
 
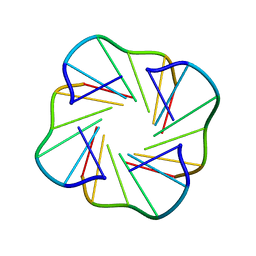 | | NMR structure of TGLGLT quadruplex | | Descriptor: | 5'-D(*TP*G LCGP*G LCGP*TP)-3' | | Authors: | Nielsen, J.T, Petersen, M. | | Deposit date: | 2006-03-15 | | Release date: | 2006-04-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structures of Lna (Locked Nucleic Acid) Modified Quadruplexes
Nucleic Acids Res., 34, 2006
|
|
6EHO
 
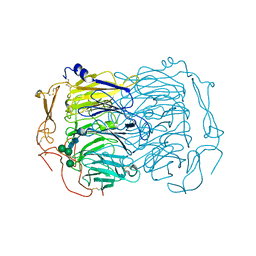 | | Dimer of the Sortilin Vps10p domain at low pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Sortilin, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Thirup, S.S, Quistgaard, E.H, Januliene, D, Andersen, J.L, Nielsen, J.A. | | Deposit date: | 2017-09-14 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Acidic Environment Induces Dimerization and Ligand Binding Site Collapse in the Vps10p Domain of Sortilin.
Structure, 25, 2017
|
|
1HV0
 
 | | DISSECTING ELECTROSTATIC INTERACTIONS AND THE PH-DEPENDENT ACTIVITY OF A FAMILY 11 GLYCOSIDASE | | Descriptor: | ENDO-1,4-BETA-XYLANASE | | Authors: | Joshi, M.D, Sidhu, G, Nielsen, J.E, Brayer, G.D, Withers, S.G, McIntosh, L.P. | | Deposit date: | 2001-01-05 | | Release date: | 2001-09-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Dissecting the electrostatic interactions and pH-dependent activity of a family 11 glycosidase.
Biochemistry, 40, 2001
|
|
7U4A
 
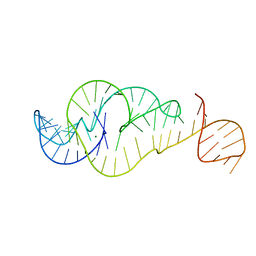 | | Crystal Structure of Zika virus xrRNA1 mutant | | Descriptor: | MAGNESIUM ION, RNA (70-MER) | | Authors: | Thompson, R.D, Carbaugh, D.L, Nielsen, J.R, Witt, C, Meganck, R.M, Rangadurai, A, Zhao, B, Bonin, J.P, Nathan, N.T, Marzluff, W.F, Frank, A.T, Lazear, H.M, Zhang, Q. | | Deposit date: | 2022-02-28 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Dynamic Basis of Xrn1 Resistance in Mosquito-borne Flavivirus RNA
To Be Published
|
|
1HV1
 
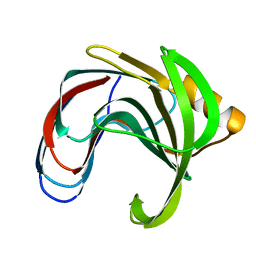 | | DISSECTING ELECTROSTATIC INTERACTIONS AND THE PH-DEPENDENT ACTIVITY OF A FAMILY 11 GLYCOSIDASE | | Descriptor: | ENDO-1,4-BETA-XYLANASE | | Authors: | Joshi, M.D, Sidhu, G, Nielsen, J.E, Brayer, G.D, Withers, S.G, McIntosh, L.P. | | Deposit date: | 2001-01-05 | | Release date: | 2001-09-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dissecting the electrostatic interactions and pH-dependent activity of a family 11 glycosidase.
Biochemistry, 40, 2001
|
|
6QUB
 
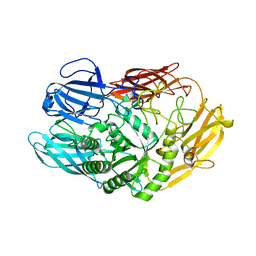 | | Truncated beta-galactosidase III from Bifidobacterium bifidum in complex with galactose | | Descriptor: | Beta-galactosidase, CALCIUM ION, beta-D-galactopyranose | | Authors: | Thirup, S.S, Nielsen, J.A, Andersen, J.L, Alsarraf, H, Blaise, M. | | Deposit date: | 2019-02-27 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Truncated beta-galactosidase III from Bifidobacterium bifidum
To Be Published
|
|
6QUC
 
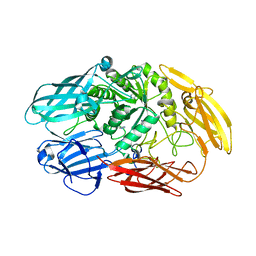 | | Truncated beta-galactosidase III from Bifidobacterium bifidum | | Descriptor: | Beta-galactosidase, CALCIUM ION, IMIDAZOLE | | Authors: | Thirup, S.S, Nielsen, J.A, Andersen, J.L, Alsarraf, H, Blaise, M. | | Deposit date: | 2019-02-27 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Truncated beta-galactosidase III from Bifidobacterium bifidum
To Be Published
|
|
6QUD
 
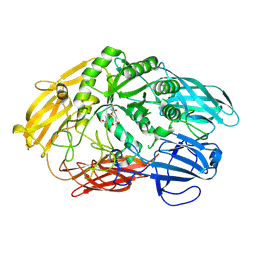 | | 2-deoxy-galactose reaction intermediate of a Truncated beta-galactosidase III from Bifidobacterium bifidum | | Descriptor: | 2-deoxy-alpha-D-galactopyranose, Beta-galactosidase, CALCIUM ION | | Authors: | Thirup, S.S, Nielsen, J.A, Andersen, J.L, Alsarraf, H, Blaise, M. | | Deposit date: | 2019-02-27 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Truncated beta-galactosidase III from Bifidobacterium bifidum
To Be Published
|
|
1NCG
 
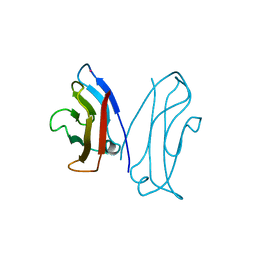 | | STRUCTURAL BASIS OF CELL-CELL ADHESION BY CADHERINS | | Descriptor: | N-CADHERIN, YTTERBIUM (III) ION | | Authors: | Shapiro, L, Fannon, A.M, Kwong, P.D, Thompson, A, Lehmann, M.S, Grubel, G, Legrand, J.-F, Als-Nielsen, J, Colman, D.R, Hendrickson, W.A. | | Deposit date: | 1995-03-23 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of cell-cell adhesion by cadherins.
Nature, 374, 1995
|
|
1NCI
 
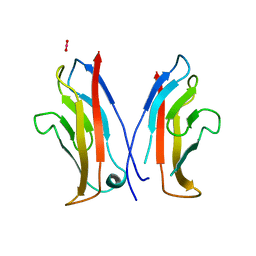 | | STRUCTURAL BASIS OF CELL-CELL ADHESION BY CADHERINS | | Descriptor: | N-CADHERIN, URANYL (VI) ION | | Authors: | Shapiro, L, Fannon, A.M, Kwong, P.D, Thompson, A, Lehmann, M.S, Grubel, G, Legrand, J.-F, Als-Nielsen, J, Colman, D.R, Hendrickson, W.A. | | Deposit date: | 1995-03-23 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of cell-cell adhesion by cadherins.
Nature, 374, 1995
|
|
1NCH
 
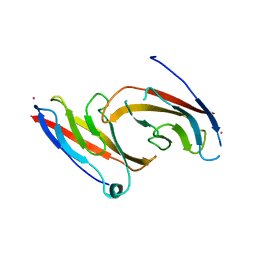 | | STRUCTURAL BASIS OF CELL-CELL ADHESION BY CADHERINS | | Descriptor: | N-CADHERIN, YTTERBIUM (III) ION | | Authors: | Shapiro, L, Fannon, A.M, Kwong, P.D, Thompson, A, Lehmann, M.S, Grubel, G, Legrand, J.-F, Als-Nielsen, J, Colman, D.R, Hendrickson, W.A. | | Deposit date: | 1995-03-23 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of cell-cell adhesion by cadherins.
Nature, 374, 1995
|
|
4CG0
 
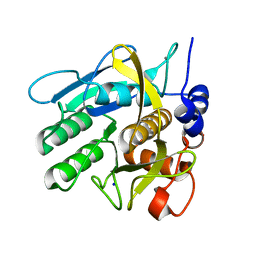 | | Savinase crystal structures for combined single crystal diffraction and powder diffraction analysis | | Descriptor: | CALCIUM ION, SODIUM ION, SUBTILISIN SAVINASE | | Authors: | Frankaer, C.G, Moroz, O.V, Turkenburg, J.P, Aspmo, S.I, Thymark, M, Friis, E.P, Stahla, K, Nielsen, J.E, Wilson, K.S, Harris, P. | | Deposit date: | 2013-11-19 | | Release date: | 2014-04-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Analysis of an Industrial Production Suspension of Bacillus Lentus Subtilisin Crystals by Powder Diffraction: A Powerful Quality-Control Tool.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4CFZ
 
 | | SAVINASE CRYSTAL STRUCTURES FOR COMBINED SINGLE CRYSTAL DIFFRACTION AND POWDER DIFFRACTION ANALYSIS | | Descriptor: | CALCIUM ION, SODIUM ION, SUBTILISIN SAVINASE, ... | | Authors: | Frankaer, C.G, Moroz, O.V, Turkenburg, J.P, Aspmo, S.I, Thymark, M, Friis, E.P, Stahla, K, Nielsen, J.E, Wilson, K.S, Harris, P. | | Deposit date: | 2013-11-19 | | Release date: | 2014-04-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Analysis of an Industrial Production Suspension of Bacillus Lentus Subtilisin Crystals by Powder Diffraction: A Powerful Quality-Control Tool.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
