3DRT
 
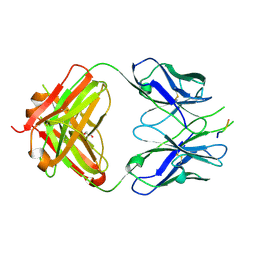 | | Crystal structure of the HIV-1 broadly neutralizing antibody 2F5 in complex with the gp41 scrambledFP-MPER scrHyb3K construct GIGAFGLLGFLAAGSKK-Ahx-K656NEQELLELDKWASLWN671 | | Descriptor: | 2F5 Fab' heavy chain, 2F5 Fab' light chain, GLYCEROL, ... | | Authors: | Julien, J.-P, Bryson, S, de la Torre, B.G, Andreu, D, Nieva, J.L, Pai, E.F. | | Deposit date: | 2008-07-11 | | Release date: | 2009-06-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural constraints imposed by the conserved fusion peptide on the HIV-1 gp41 epitope recognized by the broadly neutralizing antibody 2F5.
J.Phys.Chem.B, 113, 2009
|
|
3VZL
 
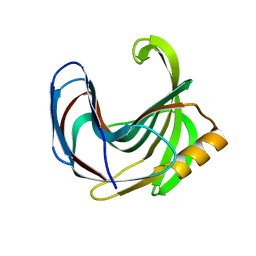 | | Crystal structure of the Bacillus circulans endo-beta-(1,4)-xylanase (BcX) N35H mutant | | Descriptor: | Endo-1,4-beta-xylanase, SULFATE ION | | Authors: | Ludwiczek, M.L, D'Angelo, I, Yalloway, G.N, Okon, M, Nielsen, J.E, Strynadka, N.C, Withers, S.G, McIntosh, L.P. | | Deposit date: | 2012-10-15 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Strategies for modulating the pH-dependent activity of a family 11 glycoside hydrolase
Biochemistry, 52, 2013
|
|
3VZN
 
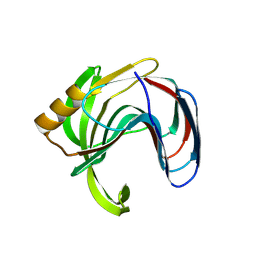 | | Crystal structure of the Bacillus circulans endo-beta-(1,4)-xylanase (BcX) N35E mutant with Glu78 covalently bonded to 2-deoxy-2-fluoro-xylobiose | | Descriptor: | Endo-1,4-beta-xylanase, SULFATE ION, beta-D-xylopyranose-(1-4)-1,5-anhydro-2-deoxy-2-fluoro-D-xylitol | | Authors: | Ludwiczek, M.L, D'Angelo, I, Yalloway, G.N, Okon, M, Nielsen, J.E, Strynadka, N.C, Withers, S.G, McIntosh, L.P. | | Deposit date: | 2012-10-15 | | Release date: | 2013-05-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Strategies for modulating the pH-dependent activity of a family 11 glycoside hydrolase
Biochemistry, 52, 2013
|
|
3EGS
 
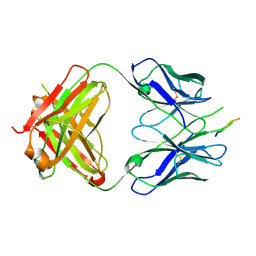 | | Crystal structure of the HIV-1 broadly neutralizing antibody 2F5 in complex with the gp41 scrambledFP-MPER scrHyb3K construct GIGAFGLLGFLAAGSKK-Ahx-K656NEQELLELDKWASLWN671 soaked in ammonium sulfate | | Descriptor: | 2F5 Fab' heavy chain, 2F5 Fab' light chain, gp41 scrFP-MPER construct | | Authors: | Julien, J.-P, Bryson, S, de la Torre, B.G, Andreu, D, Nieva, J.L, Pai, E.F. | | Deposit date: | 2008-09-11 | | Release date: | 2009-08-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural constraints imposed by the conserved fusion peptide on the HIV-1 gp41 epitope recognized by the broadly neutralizing antibody 2F5.
J.Phys.Chem.B, 113, 2009
|
|
4MVN
 
 | | Crystal structure of the staphylococcal serine protease SplA in complex with a specific phosphonate inhibitor | | Descriptor: | Serine protease splA, [(1S)-1-{[(benzyloxy)carbonyl]amino}-2-phenylethyl]phosphonic acid | | Authors: | Zdzalik, M, Burchacka, E, Niemczyk, J.S, Pustelny, K, Popowicz, G.M, Wladyka, B, Dubin, A, Potempa, J, Sienczyk, M, Dubin, G, Oleksyszyn, J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-01-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development and binding characteristics of phosphonate inhibitors of SplA protease from Staphylococcus aureus.
Protein Sci., 23, 2014
|
|
3VZJ
 
 | | Crystal structure of the Bacillus circulans endo-beta-(1,4)-xylanase (BcX) E172H mutant | | Descriptor: | Endo-1,4-beta-xylanase, SULFATE ION | | Authors: | Ludwiczek, M.L, D'Angelo, I, Yalloway, G.N, Okon, M, Nielsen, J.E, Strynadka, N.C, Withers, S.G, McIntosh, L.P. | | Deposit date: | 2012-10-14 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Strategies for modulating the pH-dependent activity of a family 11 glycoside hydrolase
Biochemistry, 52, 2013
|
|
3VZM
 
 | | Crystal structure of the Bacillus circulans endo-beta-(1,4)-xylanase (BcX) E172H mutant with Glu78 covalently bonded to 2-deoxy-2-fluoro-xylobiose | | Descriptor: | Endo-1,4-beta-xylanase, beta-D-xylopyranose-(1-4)-1,5-anhydro-2-deoxy-2-fluoro-D-xylitol | | Authors: | Ludwiczek, M.L, D'Angelo, I, Yalloway, G.N, Okon, M, Nielsen, J.E, Strynadka, N.C, Withers, S.G, McIntosh, L.P. | | Deposit date: | 2012-10-15 | | Release date: | 2013-05-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Strategies for modulating the pH-dependent activity of a family 11 glycoside hydrolase
Biochemistry, 52, 2013
|
|
3VZO
 
 | | Crystal structure of the Bacillus circulans endo-beta-(1,4)-xylanase (BcX) N35H mutant with Glu78 covalently bonded to 2-deoxy-2-fluoro-xylobiose | | Descriptor: | Endo-1,4-beta-xylanase, SULFATE ION, beta-D-xylopyranose-(1-4)-1,5-anhydro-2-deoxy-2-fluoro-D-xylitol | | Authors: | Ludwiczek, M.L, D'Angelo, I, Yalloway, G.N, Okon, M, Nielsen, J.E, Strynadka, N.C, Withers, S.G, McIntosh, L.P. | | Deposit date: | 2012-10-15 | | Release date: | 2013-05-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Strategies for modulating the pH-dependent activity of a family 11 glycoside hydrolase
Biochemistry, 52, 2013
|
|
3VZK
 
 | | Crystal structure of the Bacillus circulans endo-beta-(1,4)-xylanase (BcX) N35E mutant | | Descriptor: | Endo-1,4-beta-xylanase, SULFATE ION | | Authors: | Ludwiczek, M.L, D'Angelo, I, Yalloway, G.N, Okon, M, Nielsen, J.E, Strynadka, N.C, Withers, S.G, McIntosh, L.P. | | Deposit date: | 2012-10-14 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Strategies for modulating the pH-dependent activity of a family 11 glycoside hydrolase
Biochemistry, 52, 2013
|
|
4FMO
 
 | | Structure of the C-terminal domain of the Saccharomyces cerevisiae MUTL alpha (MLH1/PMS1) heterodimer bound to a fragment of exo1 | | Descriptor: | DNA mismatch repair protein MLH1, DNA mismatch repair protein PMS1, DNA repair peptide, ... | | Authors: | Gueneau, E, Legrand, P, Charbonnier, J.B. | | Deposit date: | 2012-06-18 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structure of the MutL alpha C-terminal domain reveals how Mlh1 contributes to Pms1 endonuclease site.
Nat.Struct.Mol.Biol., 20, 2013
|
|
3EF3
 
 | | cut-1a; NCN-Pt-Pincer-Cutinase Hybrid | | Descriptor: | (2,6-bis[(dimethylamino-kappaN)methyl]-4-{3-[(S)-ethoxy(4-nitrophenoxy)phosphoryl]propyl}phenyl-kappaC~1~)(chloro)platinum(2+), Cutinase-1 | | Authors: | Rutten, L, Mannie, J.P.B.A, Lutz, M, Gros, P. | | Deposit date: | 2008-09-08 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Solid-state structural characterization of cutinase-ECE-pincer-metal hybrids
Chemistry, 15, 2009
|
|
3ESD
 
 | | cut-2b; NCN-Pt-Pincer-Cutinase Hybrid | | Descriptor: | Cutinase 1, bromo(4-{3-[(R)-ethoxy(4-nitrophenoxy)phosphoryl]propyl}-2,6-bis[(methylsulfanyl-kappaS)methyl]phenyl-kappaC~1~)palladium(2+) | | Authors: | Rutten, L, Mannie, J.P.B.A, Lutz, M, Gros, P. | | Deposit date: | 2008-10-05 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Solid-state structural characterization of cutinase-ECE-pincer-metal hybrids
Chemistry, 15, 2009
|
|
3ESC
 
 | | cut-2a; NCN-Pt-Pincer-Cutinase Hybrid | | Descriptor: | Cutinase 1, bromo(4-{3-[(R)-ethoxy(4-nitrophenoxy)phosphoryl]propyl}-2,6-bis[(methylsulfanyl-kappaS)methyl]phenyl-kappaC~1~)palladium(2+) | | Authors: | Rutten, L, Mannie, J.P.B.A, Lutz, M, Gros, P. | | Deposit date: | 2008-10-05 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Solid-state structural characterization of cutinase-ECE-pincer-metal hybrids
Chemistry, 15, 2009
|
|
3ESA
 
 | | cut-1b; NCN-Pt-Pincer-Cutinase Hybrid | | Descriptor: | (2,6-bis[(dimethylamino-kappaN)methyl]-4-{3-[(S)-ethoxy(4-nitrophenoxy)phosphoryl]propyl}phenyl-kappaC~1~)(chloro)platinum(2+), Cutinase 1 | | Authors: | Rutten, L, Mannie, J.P.B.A, Lutz, M, Gros, P. | | Deposit date: | 2008-10-05 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Solid-state structural characterization of cutinase-ECE-pincer-metal hybrids
Chemistry, 15, 2009
|
|
3ESB
 
 | | cut-1c; NCN-Pt-Pincer-Cutinase Hybrid | | Descriptor: | (2,6-bis[(dimethylamino-kappaN)methyl]-4-{3-[(S)-ethoxy(4-nitrophenoxy)phosphoryl]propyl}phenyl-kappaC~1~)(chloro)platinum(2+), CHLORIDE ION, Cutinase 1 | | Authors: | Rutten, L, Mannie, J.P.B.A, Lutz, M, Gros, P. | | Deposit date: | 2008-10-05 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Solid-state structural characterization of cutinase-ECE-pincer-metal hybrids
Chemistry, 15, 2009
|
|
2IPI
 
 | | Crystal Structure of Aclacinomycin Oxidoreductase | | Descriptor: | Aclacinomycin oxidoreductase (AknOx), FLAVIN-ADENINE DINUCLEOTIDE, METHYL (2S,4R)-2-ETHYL-2,5,7-TRIHYDROXY-6,11-DIOXO-4-{[2,3,6-TRIDEOXY-4-O-{2,6-DIDEOXY-4-O-[(2S,6S)-6-METHYL-5-OXOTETRAHYDRO-2H-PYRAN-2-YL]-ALPHA-D-LYXO-HEXOPYRANOSYL}-3-(DIMETHYLAMINO)-D-RIBO-HEXOPYRANOSYL]OXY}-1,2,3,4,6,11-HEXAHYDROTETRACENE-1-CARBOXYLATE | | Authors: | Sultana, A, Kursula, I, Schneider, G, Alexeev, I, Niemi, J, Mantsala, P. | | Deposit date: | 2006-10-12 | | Release date: | 2007-01-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure determination by multiwavelength anomalous diffraction of aclacinomycin oxidoreductase: indications of multidomain pseudomerohedral twinning.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
4BCZ
 
 | | Monomeric Human Cu,Zn Superoxide dismutase, loops IV and VII deleted, apo form. | | Descriptor: | SUPEROXIDE DISMUTASE [CU-ZN] | | Authors: | Saraboji, K, Awad, W, Danielsson, J, Lang, L, Kurnik, M, Marklund, S.L, Oliveberg, M, Logan, D.T. | | Deposit date: | 2012-10-03 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Global Structural Motions from the Strain of a Single Hydrogen Bond.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2Y0J
 
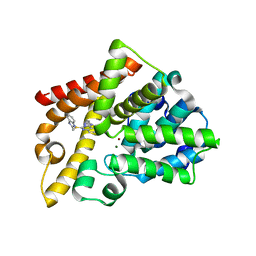 | | Triazoloquinazolines as a novel class of phosphodiesterase 10A (PDE10A) inhibitors, part 2, Lead-optimisation. | | Descriptor: | 5-(1H-BENZIMIDAZOL-2-YLMETHYLSULFANYL)-2-METHYL-[1,2,4]TRIAZOLO[1,5-C]QUINAZOLINE, CAMP AND CAMP-INHIBITED CGMP 3', 5'-CYCLIC PHOSPHODIESTERASE 10A, ... | | Authors: | Kehler, J, Ritzen, A, Langgard, M, Petersen, S.L, Christoffersen, C.T, Nielsen, J, Kilburn, J.P. | | Deposit date: | 2010-12-03 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Triazoloquinazolines as a Novel Class of Phosphodiesterase 10A (Pde10A) Inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4A3V
 
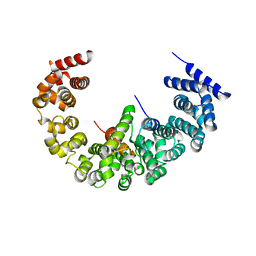 | | yeast regulatory particle proteasome assembly chaperone Hsm3 in complex with Rpt1 C-terminal fragment | | Descriptor: | 26S PROTEASE REGULATORY SUBUNIT 7 HOMOLOG, DNA MISMATCH REPAIR PROTEIN HSM3, LINKER | | Authors: | Richet, N, Barrault, M.B, Godart, C, Murciano, B, Le Tallec, B, Rousseau, E, Ledu, M.H, Charbonnier, J.B, Legrand, P, Guerois, R, Peyroche, A, Ochsenbein, F. | | Deposit date: | 2011-10-04 | | Release date: | 2012-04-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Dual Functions of the Hsm3 Protein in Chaperoning and Scaffolding Regulatory Particle Subunits During the Proteasome Assembly.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4A3T
 
 | | yeast regulatory particle proteasome assembly chaperone Hsm3 | | Descriptor: | DNA MISMATCH REPAIR PROTEIN HSM3 | | Authors: | Richet, N, Barrault, M.B, Godart, C, Murciano, B, Le Tallec, B, Rousseau, E, Ledu, M.H, Charbonnier, J.B, Legrand, P, Guerois, R, Peyroche, A, Ochsenbein, F. | | Deposit date: | 2011-10-04 | | Release date: | 2012-04-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dual Functions of the Hsm3 Protein in Chaperoning and Scaffolding Regulatory Particle Subunits During the Proteasome Assembly.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2VJT
 
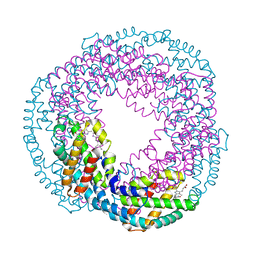 | |
1HV0
 
 | | DISSECTING ELECTROSTATIC INTERACTIONS AND THE PH-DEPENDENT ACTIVITY OF A FAMILY 11 GLYCOSIDASE | | Descriptor: | ENDO-1,4-BETA-XYLANASE | | Authors: | Joshi, M.D, Sidhu, G, Nielsen, J.E, Brayer, G.D, Withers, S.G, McIntosh, L.P. | | Deposit date: | 2001-01-05 | | Release date: | 2001-09-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Dissecting the electrostatic interactions and pH-dependent activity of a family 11 glycosidase.
Biochemistry, 40, 2001
|
|
2ZG8
 
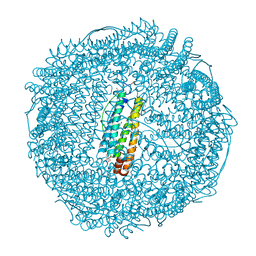 | | Crystal Structure of Pd(allyl)/apo-H49AFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, Ferritin light chain, ... | | Authors: | Abe, S, Niemeyer, J, Abe, M, Ueno, T, Hikage, T, Erker, G, Watanabe, Y. | | Deposit date: | 2008-01-18 | | Release date: | 2008-08-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Control of the coordination structure of organometallic palladium complexes in an apo-ferritin cage.
J.Am.Chem.Soc., 130, 2008
|
|
1HV1
 
 | | DISSECTING ELECTROSTATIC INTERACTIONS AND THE PH-DEPENDENT ACTIVITY OF A FAMILY 11 GLYCOSIDASE | | Descriptor: | ENDO-1,4-BETA-XYLANASE | | Authors: | Joshi, M.D, Sidhu, G, Nielsen, J.E, Brayer, G.D, Withers, S.G, McIntosh, L.P. | | Deposit date: | 2001-01-05 | | Release date: | 2001-09-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dissecting the electrostatic interactions and pH-dependent activity of a family 11 glycosidase.
Biochemistry, 40, 2001
|
|
2VJR
 
 | |
