2WP1
 
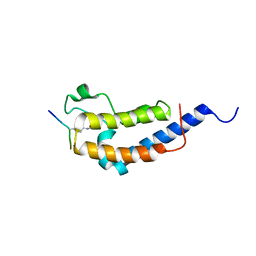 | | Structure of Brdt bromodomain 2 bound to an acetylated histone H3 peptide | | Descriptor: | BROMODOMAIN TESTIS-SPECIFIC PROTEIN, HISTONE H3 | | Authors: | Moriniere, J, Rousseaux, S, Steuerwald, U, Soler-Lopez, M, Curtet, S, Vitte, A.-L, Govin, J, Gaucher, J, Sadoul, K, Hart, D.J, Krijgsveld, J, Khochbin, S, Mueller, C.W, Petosa, C. | | Deposit date: | 2009-08-02 | | Release date: | 2009-09-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cooperative Binding of Two Acetylation Marks on a Histone Tail by a Single Bromodomain.
Nature, 461, 2009
|
|
3N6W
 
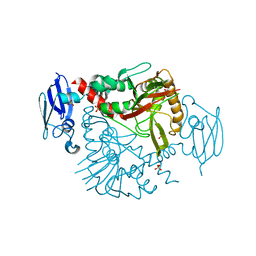 | | Crystal structure of human gamma-butyrobetaine hydroxylase | | Descriptor: | Gamma-butyrobetaine dioxygenase, SULFATE ION, ZINC ION | | Authors: | Rumnieks, J, Zeltins, A, Leonchiks, A, Kazaks, A, Kotelovica, S, Tars, K. | | Deposit date: | 2010-05-26 | | Release date: | 2010-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human gamma-butyrobetaine hydroxylase.
Biochem.Biophys.Res.Commun., 398, 2010
|
|
5HCN
 
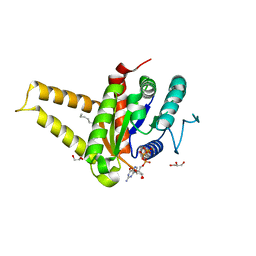 | | GPN-loop GTPase Npa3 in complex with GMPPCP | | Descriptor: | GLYCEROL, GPN-loop GTPase 1, LAURIC ACID, ... | | Authors: | Niesser, J, Wagner, F.R, Kostrewa, D, Muehlbacher, W, Cramer, P. | | Deposit date: | 2016-01-04 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of GPN-Loop GTPase Npa3 and Implications for RNA Polymerase II Assembly.
Mol.Cell.Biol., 36, 2015
|
|
6TKT
 
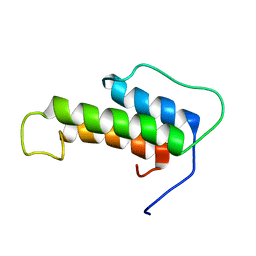 | |
2OBH
 
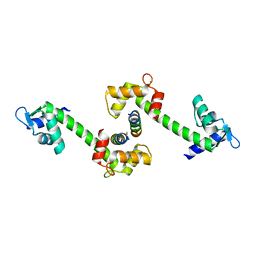 | | Centrin-XPC peptide | | Descriptor: | CALCIUM ION, Centrin-2, DNA-repair protein complementing XP-C cells | | Authors: | Charbonnier, J.B. | | Deposit date: | 2006-12-19 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural, thermodynamic, and cellular characterization of human centrin 2 interaction with xeroderma pigmentosum group C protein.
J.Mol.Biol., 373, 2007
|
|
3BII
 
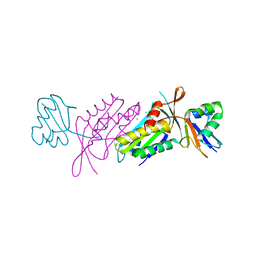 | | Crystal Structure of Activated MPT Synthase | | Descriptor: | CHLORIDE ION, Molybdopterin-converting factor subunit 1, Molybdopterin-converting factor subunit 2 | | Authors: | Daniels, J.N, Schindelin, H. | | Deposit date: | 2007-11-30 | | Release date: | 2008-02-19 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a molybdopterin synthase-precursor Z complex: insight into its sulfur transfer mechanism and its role in molybdenum cofactor deficiency.
Biochemistry, 47, 2008
|
|
4L8H
 
 | |
1KNO
 
 | |
1V02
 
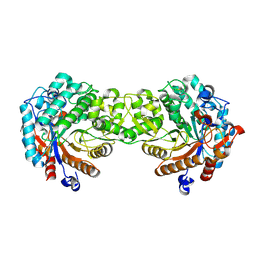 | | Crystal structure of the Sorghum bicolor dhurrinase 1 | | Descriptor: | DHURRINASE | | Authors: | Moriniere, J, Verdoucq, L, Bevan, D.R, Esen, A, Henrissat, B, Czjzek, M. | | Deposit date: | 2004-03-22 | | Release date: | 2004-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Determinants of Substrate Specificity in Family 1 Beta-Glucosidases: Novel Insights from the Crystal Structure of Sorghum Dhurrinase-1, a Plant Beta-Glucosidase with Strict Specificity, in Complex with its Natural Substrate
J.Biol.Chem., 279, 2004
|
|
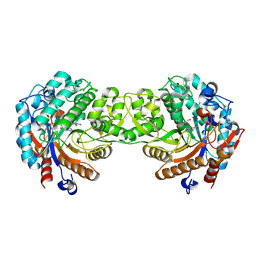
1V03
 
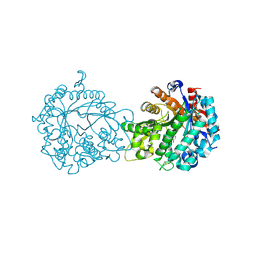 | | Crystal structure of the Sorghum bicolor dhurrinase 1 | | Descriptor: | ACETONITRILE, DHURRINASE, PHENOL, ... | | Authors: | Moriniere, J, Verdoucq, L, Bevan, D.R, Esen, A, Henrissat, B, Czjzek, M. | | Deposit date: | 2004-03-22 | | Release date: | 2004-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Determinants of Substrate Specificity in Family 1 Beta-Glucosidases: Novel Insights from the Crystal Structure of Sorghum Dhurrinase-1, a Plant Beta-Glucosidase with Strict Specificity, in Complex with its Natural Substrate
J.Biol.Chem., 279, 2004
|
|
1V08
 
 | | Crystal structure of the Zea maze beta-glucosidase-1 in complex with gluco-tetrazole | | Descriptor: | BETA-GLUCOSIDASE, NOJIRIMYCINE TETRAZOLE | | Authors: | Moriniere, J, Verdoucq, L, Bevan, D.R, Esen, A, Henrissat, B, Czjzek, M. | | Deposit date: | 2004-03-25 | | Release date: | 2004-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Determinants of Substrate Specificity in Family 1 Beta-Glucosidases: Novel Insights from the Crystal Structure of Sorghum Dhurrinase-1, a Plant Beta-Glucosidase with Strict Specificity, in Complex with its Natural Substrate
J.Biol.Chem., 279, 2004
|
|
2QIE
 
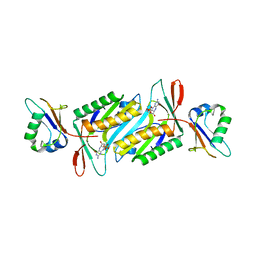 | | Staphylococcus aureus molybdopterin synthase in complex with precursor Z | | Descriptor: | (2R,4AR,5AR,11AR,12AS)-8-AMINO-2-HYDROXY-4A,5A,9,11,11A,12A-HEXAHYDRO[1,3,2]DIOXAPHOSPHININO[4',5':5,6]PYRANO[3,2-G]PTERIDINE-10,12(4H,6H)-DIONE 2-OXIDE, Molybdopterin synthase small subunit, Molybdopterin-converting factor subunit 2 | | Authors: | Daniels, J.N, Schindelin, H. | | Deposit date: | 2007-07-04 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a molybdopterin synthase-precursor Z complex: insight into its sulfur transfer mechanism and its role in molybdenum cofactor deficiency.
Biochemistry, 47, 2008
|
|
2CHK
 
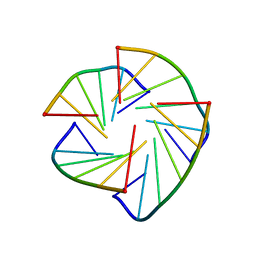 | | NMR structure of TLLLLT quadruplex | | Descriptor: | 5'-D(*T LCG LCG LCG LCGP*TP)-3' | | Authors: | Nielsen, J.T, Petersen, M. | | Deposit date: | 2006-03-15 | | Release date: | 2006-04-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structures of Lna (Locked Nucleic Acid) Modified Quadruplexes
Nucleic Acids Res., 34, 2006
|
|
2CHJ
 
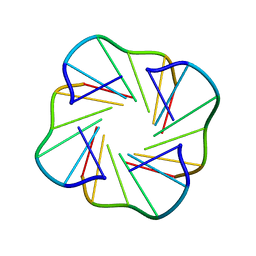 | | NMR structure of TGLGLT quadruplex | | Descriptor: | 5'-D(*TP*G LCGP*G LCGP*TP)-3' | | Authors: | Nielsen, J.T, Petersen, M. | | Deposit date: | 2006-03-15 | | Release date: | 2006-04-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structures of Lna (Locked Nucleic Acid) Modified Quadruplexes
Nucleic Acids Res., 34, 2006
|
|
3RLK
 
 | |
3RLC
 
 | |
2WCN
 
 | | Solution structure of an LNA-modified quadruplex | | Descriptor: | DNA (5'-D(*DGP*LCG*DGP*LCG*DTP*DTP*DTP *DTP*DGP*LCG*DGP*LCG)-3') | | Authors: | Nielsen, J.T, Arar, K, Petersen, M. | | Deposit date: | 2009-03-12 | | Release date: | 2009-11-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Locked Nucleic Acid Modified Quadruplex: Introducing the V4 Folding Topology.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
1OKF
 
 | | NMR structure of an alpha-L-LNA:RNA hybrid | | Descriptor: | 5'-D(*CP*ATLP*GP*AP*ATLP*AP*ATLP*GP*CP)-3', 5'-R(*GP*CP*AP*UP*AP*UP*CP*AP*GP)-3' | | Authors: | Nielsen, J.T, Stein, P.C, Petersen, M. | | Deposit date: | 2003-07-23 | | Release date: | 2003-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of an Alpha-L-Lna:RNA Hybrid: Structural Implications for Rnase H Recognition
Nucleic Acids Res., 31, 2003
|
|
2Q5W
 
 | |
4NF4
 
 | | Crystal structure of GluN1/GluN2A ligand-binding domain in complex with DCKA and glutamate | | Descriptor: | 4-hydroxy-5,7-dimethylquinoline-2-carboxylic acid, GLUTAMIC ACID, Glutamate receptor ionotropic, ... | | Authors: | Annie, J, Tajima, N, Furukawa, H. | | Deposit date: | 2013-10-30 | | Release date: | 2014-03-12 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into Competitive Antagonism in NMDA Receptors.
Neuron, 81, 2014
|
|
4E2A
 
 | |
8IQ6
 
 | | Cryo-EM structure of Latanoprost-bound prostaglandin-F2-alpha receptor-miniGq-Nb35 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Lv, X, Gao, K, Nie, J, Zhang, X, Zhang, S, Ren, Y, Li, Q, Huang, J, Liu, L, Zhang, X, Sun, X, Zhang, W, Liu, X. | | Deposit date: | 2023-03-15 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of human prostaglandin F 2 alpha receptor reveal the mechanism of ligand and G protein selectivity.
Nat Commun, 14, 2023
|
|
8IQ4
 
 | | Cryo-EM structure of Carboprost-bound prostaglandin-F2-alpha receptor-miniGq-Nb35 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Lv, X, Gao, K, Nie, J, Zhang, X, Zhang, S, Ren, Y, Li, Q, Huang, J, Liu, L, Zhang, X, Sun, X, Zhang, W, Liu, X. | | Deposit date: | 2023-03-15 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures of human prostaglandin F 2 alpha receptor reveal the mechanism of ligand and G protein selectivity.
Nat Commun, 14, 2023
|
|
8J2M
 
 | | The truncated rice Na+/H+ antiporter SOS1 (1-976) in a constitutively active state | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate, Na+/H+ antiporter | | Authors: | Zhang, X.Y, Tang, L.H, Zhang, C.R, Nie, J.W. | | Deposit date: | 2023-04-14 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure and activation mechanism of the rice Salt Overly Sensitive 1 (SOS1) Na + /H + antiporter.
Nat.Plants, 9, 2023
|
|
8IWO
 
 | | The rice Na+/H+ antiporter SOS1 in an auto-inhibited state | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate, OsSOS1 | | Authors: | Zhang, X.Y, Tang, L.H, Zhang, C.R, Nie, J.W. | | Deposit date: | 2023-03-30 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structure and activation mechanism of the rice Salt Overly Sensitive 1 (SOS1) Na + /H + antiporter.
Nat.Plants, 9, 2023
|
|
