2GF0
 
 | | The crystal structure of the human DiRas1 GTPase in the inactive GDP bound state | | Descriptor: | GTP-binding protein Di-Ras1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Turnbull, A.P, Papagrigoriou, E, Yang, X, Schoch, G, Elkins, J, Gileadi, O, Salah, E, Bray, J, Wen-Hwa, L, Fedorov, O, Niesen, F.E, von Delft, F, Weigelt, J, Edwards, A, Arrowsmith, C, Sundstrom, M, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-03-21 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of the human DiRas1 GTPase in the inactive GDP bound state
To be Published
|
|
1CF2
 
 | | THREE-DIMENSIONAL STRUCTURE OF D-GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE FROM THE HYPERTHERMOPHILIC ARCHAEON METHANOTHERMUS FERVIDUS | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTEIN (GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE), SULFATE ION | | Authors: | Charron, C, Talfournier, F, Isuppov, M.N, Branlant, G, Littlechild, J.A, Vitoux, B, Aubry, A. | | Deposit date: | 1999-03-24 | | Release date: | 2000-03-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallization and preliminary X-ray diffraction studies of D-glyceraldehyde-3-phosphate dehydrogenase from the hyperthermophilic archaeon Methanothermus fervidus.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1SAW
 
 | | X-ray structure of homo sapiens protein FLJ36880 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, hypothetical protein FLJ36880 | | Authors: | Manjasetty, B.A, Niesen, F.H, Delbrueck, H, Goetz, F, Sievert, V, Buessow, K, Behlke, J, Heinemann, U. | | Deposit date: | 2004-02-09 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structure of fumarylacetoacetate hydrolase family member Homo sapiens FLJ36880.
Biol.Chem., 385, 2004
|
|
2VD5
 
 | | Structure of Human Myotonic Dystrophy Protein Kinase in Complex with the Bisindoylmaleide inhibitor BIM VIII | | Descriptor: | 3-[1-(3-AMINOPROPYL)-1H-INDOL-3-YL]-4-(1-METHYL-1H-INDOL-3-YL)-1H-PYRROLE-2,5-DIONE, DMPK PROTEIN | | Authors: | Pike, A.C.W, Amos, A, Elkins, J, Bullock, A, Guo, K, Fedorov, O, Bunkoczi, G, Filippakopoulos, P, Pilka, E.S, Ugochukwu, E, Umeano, C, Niesen, F, Sundstrom, M, Weigelt, J, Edwards, A, Arrowsmith, C.H, von Delft, F, Knapp, S. | | Deposit date: | 2007-09-30 | | Release date: | 2007-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Dystrophia Myotonica Protein Kinase.
Protein Sci., 18, 2009
|
|
2WM3
 
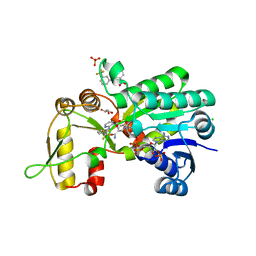 | | Crystal structure of NmrA-like family domain containing protein 1 in complex with niflumic acid | | Descriptor: | 2-{[3-(TRIFLUOROMETHYL)PHENYL]AMINO}NICOTINIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Bhatia, C, Yue, W.W, Niesen, F, Pilka, E, Ugochukwu, E, Savitsky, P, Hozjan, V, Roos, A.K, Filippakopoulos, P, von Delft, F, Heightman, T, Arrowsmith, C, Weigelt, J, Edwards, A, Bountra, C, Oppermann, U. | | Deposit date: | 2009-06-29 | | Release date: | 2009-08-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Nmra-Like Family Domain Containing Protein 1 in Complex with Niflumic Acid
To be Published
|
|
1Z57
 
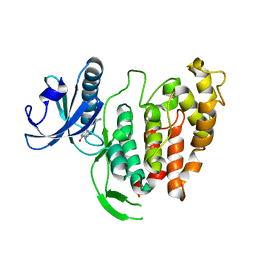 | | Crystal structure of human CLK1 in complex with 10Z-Hymenialdisine | | Descriptor: | DEBROMOHYMENIALDISINE, Dual specificity protein kinase CLK1 | | Authors: | Debreczeni, J, Das, S, Knapp, S, Bullock, A, Guo, K, Amos, A, Fedorov, O, Edwards, A, Sundstrom, M, von Delft, F, Niesen, F.H, Ball, L, Sobott, F, Arrowsmith, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-03-17 | | Release date: | 2005-04-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Kinase domain insertions define distinct roles of CLK kinases in SR protein phosphorylation.
Structure, 17, 2009
|
|
2AR7
 
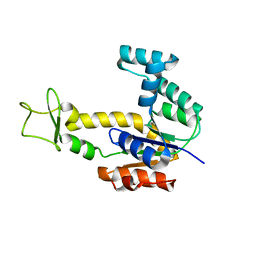 | | Crystal structure of human adenylate kinase 4, AK4 | | Descriptor: | Adenylate kinase 4 | | Authors: | Filippakopoulos, P, Turnbull, A.P, Fedorov, O, Weigelt, J, Bunkoczi, G, Ugochukwu, E, Debreczeni, J, Niesen, F, von Delft, F, Edwards, A, Arrowsmith, C, Sundstrom, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-08-19 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of human adenylate kinase 4, AK4
To be Published
|
|
6GWX
 
 | | Stabilising and Understanding a Miniprotein by Rational Design. | | Descriptor: | Optimised PPa-TYR | | Authors: | Porter Goff, K.L, Williams, C, Baker, E.G, Nicol, D, Samphire, J.L, Zieleniewski, F.L, Crump, M.P, Woolfson, D.N. | | Deposit date: | 2018-06-26 | | Release date: | 2019-07-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Stabilizing and Understanding a Miniprotein by Rational Redesign.
Biochemistry, 58, 2019
|
|
1O94
 
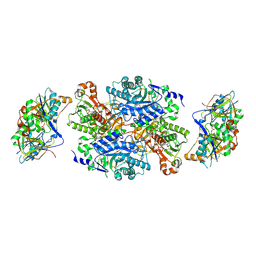 | | Ternary complex between trimethylamine dehydrogenase and electron transferring flavoprotein | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ELECTRON TRANSFER FLAVOPROTEIN ALPHA-SUBUNIT, ... | | Authors: | Leys, D, Basran, J, Talfournier, F, Sutcliffe, M.J, Scrutton, N.S. | | Deposit date: | 2002-12-11 | | Release date: | 2003-02-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Extensive Conformational Sampling in a Ternary Electron Transfer Complex.
Nat.Struct.Biol., 10, 2003
|
|
1O96
 
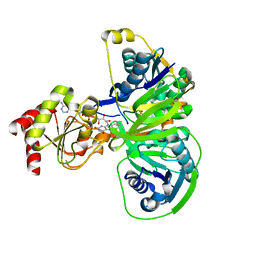 | | Structure of electron transferring flavoprotein for Methylophilus methylotrophus. | | Descriptor: | ADENOSINE MONOPHOSPHATE, ELECTRON TRANSFERRING FLAVOPROTEIN ALPHA-SUBUNIT, ELECTRON TRANSFERRING FLAVOPROTEIN BETA-SUBUNIT, ... | | Authors: | Leys, D, Basran, J, Talfournier, F, Sutcliffe, M.J, Scrutton, N.S. | | Deposit date: | 2002-12-11 | | Release date: | 2003-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Extensive Conformational Sampling in a Ternary Electron Transfer Complex.
Nat.Struct.Biol., 10, 2003
|
|
4LRS
 
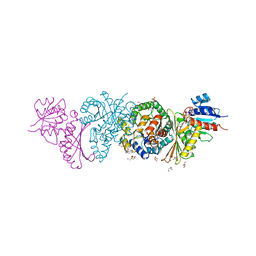 | | Crystal and solution structures of the bifunctional enzyme (Aldolase/Aldehyde dehydrogenase) from Thermomonospora curvata, reveal a cofactor-binding domain motion during NAD+ and CoA accommodation whithin the shared cofactor-binding site | | Descriptor: | 4-hydroxy-2-oxovalerate aldolase, Acetaldehyde dehydrogenase, CHLORIDE ION, ... | | Authors: | Fischer, B, Branlant, G, Talfournier, F, Gruez, A. | | Deposit date: | 2013-07-20 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal and solution structures of the bifunctional enzyme (Aldolase/Aldehyde dehydrogenase) from Thermomonospora curvata, reveal a cofactor-binding domain motion during NAD+ and CoA accommodation whithin the shared cofactor-binding site
To be Published
|
|
4LRT
 
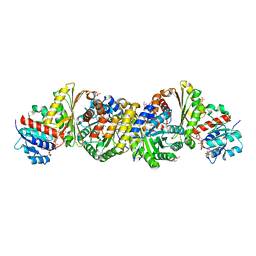 | | Crystal and solution structures of the bifunctional enzyme (Aldolase/Aldehyde dehydrogenase) from Thermomonospora curvata, reveal a cofactor-binding domain motion during NAD+ and CoA accommodation whithin the shared cofactor-binding site | | Descriptor: | 4-hydroxy-2-oxovalerate aldolase, Acetaldehyde dehydrogenase, COENZYME A, ... | | Authors: | Fischer, B, Branlant, G, Talfournier, F, Gruez, A. | | Deposit date: | 2013-07-20 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal and solution structures of the bifunctional enzyme (Aldolase/Aldehyde dehydrogenase) from Thermomonospora curvata, reveal a cofactor-binding domain motion during NAD+ and CoA accommodation whithin the shared cofactor-binding site
To be Published
|
|
2GJT
 
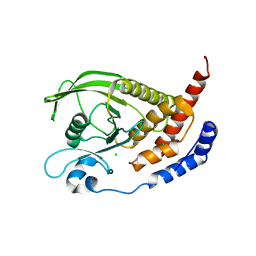 | | Crystal structure of the human receptor phosphatase PTPRO | | Descriptor: | CHLORIDE ION, Receptor-type tyrosine-protein phosphatase PTPRO | | Authors: | Barr, A, Ugochukwu, E, Eswaran, J, Das, S, Niesen, F, Savitsky, P, Turnbull, A, Sundstrom, M, Arrowsmith, C, Edwards, A, Weigelt, J, von Delft, F, Papagrigoriou, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-03-31 | | Release date: | 2006-05-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Large-scale structural analysis of the classical human protein tyrosine phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
2W98
 
 | | CRYSTAL STRUCTURE OF HUMAN ZINC-BINDING ALCOHOL DEHYDROGENASE 1 (ZADH1) IN TERNARY COMPLEX WITH NADP AND PHENYLBUTAZONE | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 4-BUTYL-1,2-DIPHENYL-PYRAZOLIDINE-3,5-DIONE, CHLORIDE ION, ... | | Authors: | Shafqat, N, Yue, W.W, Muniz, J, Picaud, S, Niesen, F, Arrowsmith, C, Weigelt, J, Edwards, A, Bountra, C, Oppermann, U. | | Deposit date: | 2009-01-22 | | Release date: | 2009-04-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Human Zinc-Binding Alcohol Dehydrogenase 1
To be Published
|
|
2WMD
 
 | | Crystal structure of NmrA-like family domain containing protein 1 in complex with NADP and 2-(4-chloro-phenylamino)-nicotinic acid | | Descriptor: | 2-(4-CHLORO-PHENYLAMINO)-NICOTINIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NMRA-LIKE FAMILY DOMAIN CONTAINING PROTEIN 1 | | Authors: | Bhatia, C, Pilka, E, Niesen, F, Yue, W.W, Ugochukwu, E, Savitsky, P, Hozjan, V, Roos, A.K, Filippakopoulos, P, von Delft, F, Heightman, T, Arrowsmith, C, Weigelt, J, Edwards, A, Bountra, C, Oppermann, U. | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Nmra-Like Family Domain Containing Protein 1
To be Published
|
|
2W4Q
 
 | | Crystal structure of human zinc-binding alcohol dehydrogenase 1 (ZADH1) in ternary complex with NADP and 18beta-glycyrrhetinic acid | | Descriptor: | (3BETA,5BETA,14BETA)-3-HYDROXY-11-OXOOLEAN-12-EN-29-OIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROSTAGLANDIN REDUCTASE 2, ... | | Authors: | Shafqat, N, Yue, W.W, Ugochukwu, E, Picaud, S, Niesen, F, Arrowsmith, C, Weigelt, J, Edwards, A, Bountra, C, Oppermann, U. | | Deposit date: | 2008-11-30 | | Release date: | 2009-01-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human Zinc-Binding Alcohol Dehydrogenase 1
To be Published
|
|
4H5B
 
 | | Crystal Structure of DR_1245 from Deinococcus radiodurans | | Descriptor: | BROMIDE ION, DR_1245 protein, GLYCEROL, ... | | Authors: | Norais, C, Servant, P, Bouthier-de-la-Tour, C, Coureux, P.D, Ithurbide, S, Vannier, F, Guerin, P, Dulberger, C.L, Satyshur, K.A, Keck, J.L, Armengaud, J, Cox, M.M, Sommer, S. | | Deposit date: | 2012-09-18 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Deinococcus radiodurans DR1245 Protein, a DdrB Partner Homologous to YbjN Proteins and Reminiscent of Type III Secretion System Chaperones.
Plos One, 8, 2013
|
|
1O97
 
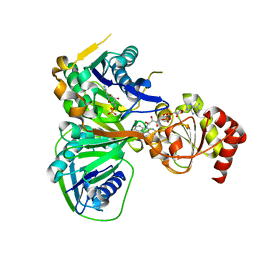 | | Structure of electron transferring flavoprotein from Methylophilus methylotrophus, recognition loop removed by limited proteolysis | | Descriptor: | ADENOSINE MONOPHOSPHATE, ELECTRON TRANSFERRING FLAVOPROTEIN ALPHA-SUBUNIT, ELECTRON TRANSFERRING FLAVOPROTEIN BETA-SUBUNIT, ... | | Authors: | Leys, D, Basran, J, Talfournier, F, Sutcliffe, M.J, Scrutton, N.S. | | Deposit date: | 2002-12-11 | | Release date: | 2003-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Extensive Conformational Sampling in a Ternary Electron Transfer Complex.
Nat.Struct.Biol., 10, 2003
|
|
1S13
 
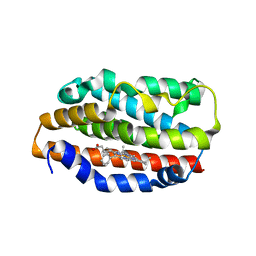 | | Human Heme Oxygenase Oxidatition of alpha- and gamma-meso-Phenylhemes | | Descriptor: | 2-PHENYLHEME, Heme oxygenase 1 | | Authors: | Wang, J, Niemevz, F, Lad, L, Buldain, G, Poulos, T.L, Ortiz de Montellano, P.R. | | Deposit date: | 2004-01-05 | | Release date: | 2004-08-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Human heme oxygenase oxidation of 5- and 15-phenylhemes.
J.Biol.Chem., 279, 2004
|
|
2QEP
 
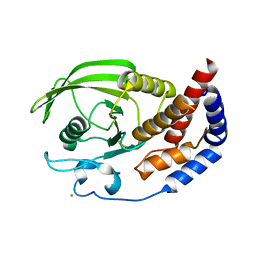 | | Crystal structure of the D1 domain of PTPRN2 (IA2beta) | | Descriptor: | CHLORIDE ION, Receptor-type tyrosine-protein phosphatase N2 | | Authors: | Ugochukwu, E, Barr, A, Alfano, I, Berridge, G, Burgess-Brown, N, Das, S, Fedorov, O, King, O, Niesen, F, Watt, S, Savitsky, P, Salah, E, Pike, A.C.W, Bunkoczi, G, von Delft, F, Sundstrom, M, Edwards, A, Arrowsmith, C.H, Weigelt, J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-26 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Large-scale structural analysis of the classical human protein tyrosine phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
3B7O
 
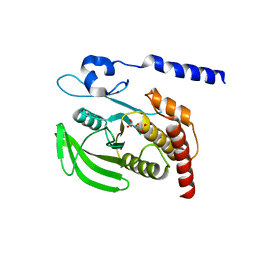 | | Crystal structure of the human tyrosine phosphatase SHP2 (PTPN11) with an accessible active site | | Descriptor: | D-MALATE, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Ugochukwu, E, Barr, A, Patel, A, King, O, Niesen, F, Salah, E, Savitsky, P, Pilka, E.S, Elkins, J, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-31 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Large-scale structural analysis of the classical human protein tyrosine phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
3DTK
 
 | | Crystal structure of the IRRE protein, a central regulator of DNA damage repair in deinococcaceae | | Descriptor: | CHLORIDE ION, IRRE PROTEIN, ZINC ION | | Authors: | Vujicic-Zagar, A, Dulermo, R, Le Gorrec, M, Vannier, F, Servant, P, Sommer, S, de Groot, A, Serre, L. | | Deposit date: | 2008-07-15 | | Release date: | 2009-02-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Crystal structure of the IrrE protein, a central regulator of DNA damage repair in deinococcaceae
J.Mol.Biol., 386, 2009
|
|
3DTI
 
 | | Crystal structure of the IRRE protein, a central regulator of DNA damage repair in deinococcaceae | | Descriptor: | IRRE protein, ZINC ION | | Authors: | Vujicic-Zagar, A, Dulermo, R, Le Gorrec, M, Vannier, F, Servant, P, Sommer, S, De Groot, A, Serre, L. | | Deposit date: | 2008-07-15 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the IrrE protein, a central regulator of DNA damage repair in deinococcaceae
J.Mol.Biol., 386, 2009
|
|
3DTE
 
 | | Crystal structure of the IRRE protein, a central regulator of DNA damage repair in deinococcaceae | | Descriptor: | IrrE protein | | Authors: | Vujicic-Zagar, A, Dulermo, R, Le Gorrec, M, Vannier, F, Servant, P, Sommer, S, De Groot, A, Serre, L. | | Deposit date: | 2008-07-15 | | Release date: | 2009-02-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the IrrE protein, a central regulator of DNA damage repair in deinococcaceae
J.Mol.Biol., 386, 2009
|
|
1JYR
 
 | | Xray Structure of Grb2 SH2 Domain Complexed with a Phosphorylated Peptide | | Descriptor: | GROWTH FACTOR RECEPTOR-BOUND PROTEIN 2, peptide: PSpYVNVQN | | Authors: | Nioche, P, Liu, W.-Q, Broutin, I, Charbonnier, F, Latreille, M.-T, Vidal, M, Roques, B, Garbay, C, Ducruix, A. | | Deposit date: | 2001-09-13 | | Release date: | 2002-03-13 | | Last modified: | 2018-10-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of the SH2 domain of Grb2: highlight on the binding of a new high-affinity inhibitor.
J.Mol.Biol., 315, 2002
|
|
