6P4S
 
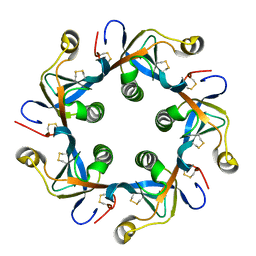 | | Salmonella typhi PltB Homopentamer T65I Mutant | | Descriptor: | Putative pertussis-like toxin subunit | | Authors: | Nguyen, T, Milano, S.K, Hillpot, E.C, Yang, Y.A, Song, J. | | Deposit date: | 2019-05-28 | | Release date: | 2020-03-25 | | Last modified: | 2020-08-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Salmonella Typhoid Toxin PltB Subunit and Its Non-typhoidal Salmonella Ortholog Confer Differential Host Adaptation and Virulence.
Cell Host Microbe, 27, 2020
|
|
6P4P
 
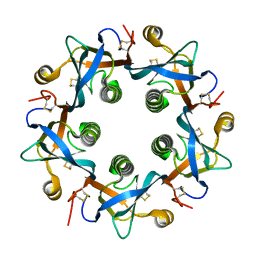 | | Salmonella typhi PltB Homopentamer N29K Mutant | | Descriptor: | Putative pertussis-like toxin subunit | | Authors: | Nguyen, T, Milano, S.K, Hillpot, E.C, Yang, Y.A, Song, J. | | Deposit date: | 2019-05-28 | | Release date: | 2020-03-25 | | Last modified: | 2020-08-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Salmonella Typhoid Toxin PltB Subunit and Its Non-typhoidal Salmonella Ortholog Confer Differential Host Adaptation and Virulence.
Cell Host Microbe, 27, 2020
|
|
3U3G
 
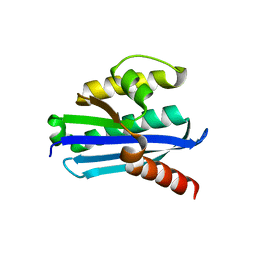 | | Structure of LC11-RNase H1 Isolated from Compost by Metagenomic Approach: Insight into the Structural Bases for Unusual Enzymatic Properties of Sto-RNase H1 | | Descriptor: | CHLORIDE ION, Ribonuclease H, UNKNOWN LIGAND | | Authors: | Nguyen, T.N, Angkawidjaja, C, Kanaya, E, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2011-10-05 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Activity, stability, and structure of metagenome-derived LC11-RNase H1, a homolog of Sulfolobus tokodaii RNase H1
Protein Sci., 21, 2012
|
|
5FWZ
 
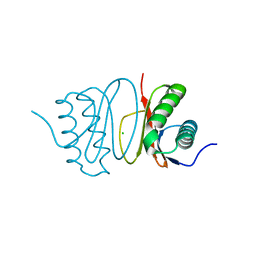 | | Fasciola hepatica calcium binding protein FhCaBP2: Structure of the dynein light chain-like domain. P41212 mercury derivative. | | Descriptor: | CALCIUM BINDING PROTEIN, CHLORIDE ION, MERCURY (II) ION | | Authors: | Nguyen, T.H, Thomas, C.M, Timson, D.J, van Raaij, M.J. | | Deposit date: | 2016-02-22 | | Release date: | 2016-04-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fasciola hepatica calcium-binding protein FhCaBP2: structure of the dynein light chain-like domain.
Parasitol. Res., 115, 2016
|
|
5FX0
 
 | |
7KM2
 
 | | Dodecameric Structure of the Chlamydia trachomatis Flavin Prenyltransferase UbiX Ortholog CT220 with FMN | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavin prenyltransferase UbiX | | Authors: | Nguyen, T, Nicely, N.I, Belaia-Martiniouk, A, Dunne, A.P, McCafferty, D.G. | | Deposit date: | 2020-11-02 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Functional and structural validation of CT220 as the UbiX-like flavin prenyltransferase from Chlamydial menaquinone biosynthesis
To be published
|
|
7KM3
 
 | | Dodecameric Structure of the Chlamydia trachomatis Flavin Prenyltransferase UbiX Ortholog CT220 with FMN and DMAP | | Descriptor: | Dimethylallyl monophosphate, FLAVIN MONONUCLEOTIDE, Flavin prenyltransferase UbiX | | Authors: | Nguyen, T, Nicely, N.I, Belaia-Martiniouk, A, Dunne, A.P, McCafferty, D.G. | | Deposit date: | 2020-11-02 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Functional and structural validation of CT220 as the UbiX-like flavin prenyltransferase from Chlamydial menaquinone biosynthesis
To be published
|
|
4H8K
 
 | | Crystal structure of LC11-RNase H1 in complex with RNA/DNA hybrid | | Descriptor: | DNA (5'-D(*GP*GP*AP*AP*TP*CP*AP*GP*GP*TP*GP*TP*CP*G)-3'), RNA (5'-R(*CP*GP*AP*CP*AP*CP*CP*UP*GP*AP*UP*UP*CP*C)-3'), Ribonuclease H | | Authors: | Nguyen, T.N, You, D.J, Matsumoto, H, Kanaya, E, Kanaya, S. | | Deposit date: | 2012-09-23 | | Release date: | 2013-09-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of metagenome-derived LC11-RNase H1 in complex with RNA/DNA hybrid
J.Struct.Biol., 182, 2013
|
|
4IBN
 
 | | Crystal structure of LC9-RNase H1, a type 1 RNase H with the type 2 active-site motif | | Descriptor: | Ribonuclease H | | Authors: | Nguyen, T.-N, You, D.-J, Kanaya, E, Koga, Y, Kanaya, S. | | Deposit date: | 2012-12-09 | | Release date: | 2013-03-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure of metagenome-derived LC9-RNase H1 with atypical DEDN active site motif
Febs Lett., 587, 2013
|
|
4BGD
 
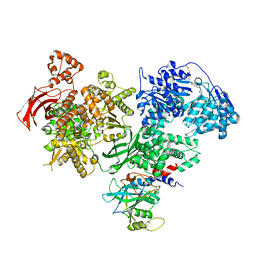 | | Crystal structure of Brr2 in complex with the Jab1/MPN domain of Prp8 | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Nguyen, T.H.D, Li, J, Nagai, K. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of Brr2-Prp8 Interactions and Implications for U5 Snrnp Biogenesis and the Spliceosome Active Site
Structure, 21, 2013
|
|
7K7I
 
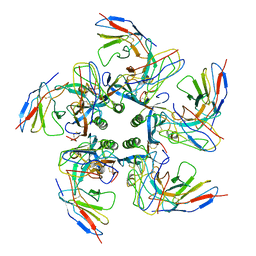 | | Density-fitted Model Structure of Antibody Variable Domains of TyTx4 in Complex with PltB pentamer of Typhoid Toxin | | Descriptor: | Fab Heavy Chain Variable Domain, Fab Light Chain Variable Domain, Putative pertussis-like toxin subunit | | Authors: | Nguyen, T, Feathers, J.R, Fromme, J.C, Song, J. | | Deposit date: | 2020-09-22 | | Release date: | 2021-09-01 | | Last modified: | 2021-09-22 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | The structural basis of Salmonella A 2 B 5 toxin neutralization by antibodies targeting the glycan-receptor binding subunits.
Cell Rep, 36, 2021
|
|
7K7H
 
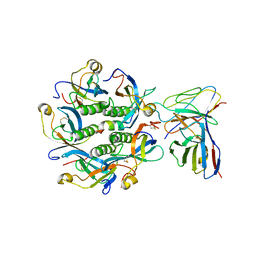 | | Density-fitted Model Structure of Antibody Variable Domains of TyTx1 in Complex with PltB pentamer of Typhoid Toxin | | Descriptor: | Fab Heavy Chain Variable Domain, Fab Light Chain Variable Domain, Pertussis like toxin subunit B, ... | | Authors: | Nguyen, T, Feathers, J.R, Fromme, J.C, Song, J. | | Deposit date: | 2020-09-22 | | Release date: | 2021-09-01 | | Last modified: | 2021-09-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The structural basis of Salmonella A 2 B 5 toxin neutralization by antibodies targeting the glycan-receptor binding subunits.
Cell Rep, 36, 2021
|
|
7KU0
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 138 (yellow) structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KU3
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 141 (cyan) structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KU1
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 139 (green) structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KTZ
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 131 (purple) structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KTY
 
 | | Data clustering and dynamics of chymotrypsinogen average structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Shi, W, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KU2
 
 | | Data clustering and dynamics of chymotrypsinogen clulster 140 (structure) | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.185 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KM4
 
 | |
7CLS
 
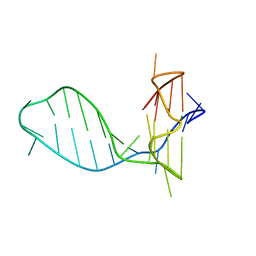 | |
1HA9
 
 | | SOLUTION STRUCTURE OF THE SQUASH TRYPSIN INHIBITOR MCoTI-II, NMR, 30 STRUCTURES. | | Descriptor: | TRYPSIN INHIBITOR II | | Authors: | Heitz, A, Hernandez, J.-F, Gagnon, J, Hong, T.T, Pham, T.T.C, Nguyen, T.M, Le-Nguyen, D, Chiche, L. | | Deposit date: | 2001-04-02 | | Release date: | 2001-04-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Squash Trypsin Inhibitor Mcoti-II. A New Family for Cyclic Knottins
Biochemistry, 40, 2001
|
|
1C4R
 
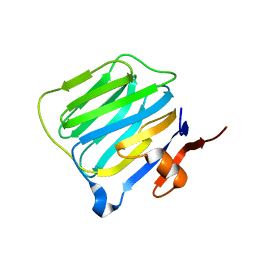 | | THE STRUCTURE OF THE LIGAND-BINDING DOMAIN OF NEUREXIN 1BETA: REGULATION OF LNS DOMAIN FUNCTION BY ALTERNATIVE SPLICING | | Descriptor: | NEUREXIN-I BETA | | Authors: | Rudenko, G, Nguyen, T, Chelliah, Y, Sudhof, T.C, Deisenhofer, J. | | Deposit date: | 1999-09-28 | | Release date: | 2000-10-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of the ligand-binding domain of neurexin Ibeta: regulation of LNS domain function by alternative splicing.
Cell(Cambridge,Mass.), 99, 1999
|
|
5K5T
 
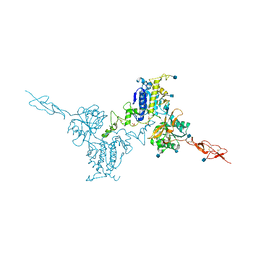 | | Crystal structure of the inactive form of human calcium-sensing receptor extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Extracellular calcium-sensing receptor, ... | | Authors: | Geng, Y, Mosyak, L, Kurinov, I, Zuo, H, Sturchler, E, Cheng, T.C, Subramanyam, P, Brown, A.P, Brennan, S.C, Mun, H.-C, Bush, M, Chen, Y, Nguyen, T, Cao, B, Chang, D, Quick, M, Conigrave, A, Colecraft, H.M, McDonald, P, Fan, Q.R. | | Deposit date: | 2016-05-23 | | Release date: | 2016-08-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural mechanism of ligand activation in human calcium-sensing receptor.
Elife, 5, 2016
|
|
5K5S
 
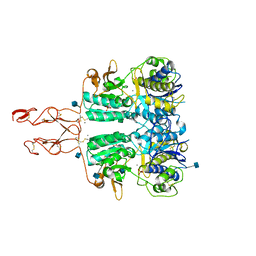 | | Crystal structure of the active form of human calcium-sensing receptor extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Extracellular calcium-sensing receptor, ... | | Authors: | Geng, Y, Mosyak, L, Kurinov, I, Zuo, H, Sturchler, E, Cheng, T.C, Subramanyam, P, Brown, A.P, Brennan, S.C, Mun, H.-C, Bush, M, Chen, Y, Nguyen, T, Cao, B, Chang, D, Quick, M, Conigrave, A, Colecraft, H.M, McDonald, P, Fan, Q.R. | | Deposit date: | 2016-05-23 | | Release date: | 2016-08-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural mechanism of ligand activation in human calcium-sensing receptor.
Elife, 5, 2016
|
|
8SZJ
 
 | | Human glutaminase C (Y466W) with L-Gln and Pi, filamentous form | | Descriptor: | GLUTAMINE, Glutaminase kidney isoform, mitochondrial, ... | | Authors: | Feng, S, Aplin, C, Nguyen, T.-T.T, Milano, S.K, Cerione, R.A. | | Deposit date: | 2023-05-29 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Filament formation drives catalysis by glutaminase enzymes important in cancer progression.
Nat Commun, 15, 2024
|
|
