1NHA
 
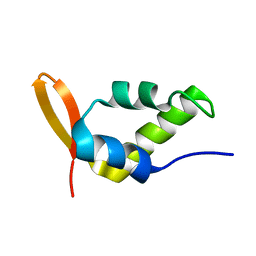 | | Solution Structure of the Carboxyl-Terminal Domain of RAP74 and NMR Characterization of the FCP-Binding Sites of RAP74 and CTD of RAP74, the subunit of Human TFIIF | | Descriptor: | Transcription initiation factor IIF, alpha subunit | | Authors: | Nguyen, B.D, Chen, H.T, Kobor, M.S, Greenblatt, J, Legault, P, Omichinski, J.G. | | Deposit date: | 2002-12-19 | | Release date: | 2003-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Carboxyl-Terminal Domain of RAP74 and NMR Characterization of the FCP1-Binding Sites of RAP74 and Human TFIIB.
Biochemistry, 42, 2003
|
|
8E7I
 
 | |
8E7H
 
 | |
8E7E
 
 | |
8E7J
 
 | |
8E7D
 
 | |
1ONV
 
 | | NMR Structure of a Complex Containing the TFIIF Subunit RAP74 and the RNAP II CTD Phosphatase FCP1 | | Descriptor: | Transcription initiation factor IIF, alpha subunit, serine phosphatase FCP1a | | Authors: | Nguyen, B.D, Abbott, K.L, Potempa, K, Kobor, M.S, Archambault, J, Greenblatt, J, Legault, P, Omichinski, J.G. | | Deposit date: | 2003-03-02 | | Release date: | 2003-05-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of a Complex Containing the TFIIF Subunit RAP74 and the RNA polymerase II carboxyl-terminal domain phosphatase FCP1
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
8TDN
 
 | |
8G9R
 
 | |
8TDO
 
 | |
8GBR
 
 | |
1M6F
 
 | | Strong Binding in the DNA Minor Groove by an Aromatic Diamidine With a Shape That Does Not Match the Curvature of the Groove | | Descriptor: | 3-[C-[N'-(3-CARBAMIMIDOYL-BENZYLIDENIUM)-HYDRAZINO]-[[AMINOMETHYLIDENE]AMINIUM]-IMINOMETHYL]-BENZAMIDINIUM, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Nguyen, B, Lee, M.P.H, Hamelberg, D, Joubert, A, Bailly, C, Brun, R, Neidle, S, Wilson, W.D. | | Deposit date: | 2002-07-16 | | Release date: | 2002-11-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Strong Binding in the DNA Minor Groove by an Aromatic Diamidine With a Shape That Does Not Match the Curvature of the Groove
J.Am.Chem.Soc., 124, 2002
|
|
1IA8
 
 | | THE 1.7 A CRYSTAL STRUCTURE OF HUMAN CELL CYCLE CHECKPOINT KINASE CHK1 | | Descriptor: | CHK1 CHECKPOINT KINASE, SULFATE ION | | Authors: | Chen, P, Luo, C, Deng, Y, Ryan, K, Register, J, Margosiak, S, Tempczyk-Russell, A, Nguyen, B, Myers, P, Lundgren, K, Chen Kan, C.-C, O'Connor, P.M. | | Deposit date: | 2001-03-22 | | Release date: | 2001-04-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The 1.7 A crystal structure of human cell cycle checkpoint kinase Chk1: implications for Chk1 regulation.
Cell(Cambridge,Mass.), 100, 2000
|
|
1Y5O
 
 | | NMR structure of the amino-terminal domain from the Tfb1 subunit of yeast TFIIH | | Descriptor: | RNA polymerase II transcription factor B 73 kDa subunit | | Authors: | Di Lello, P, Nguyen, B.D, Jones, T.N, Potempa, K, Kobor, M.S, Legault, P, Omichinski, J.G. | | Deposit date: | 2004-12-02 | | Release date: | 2005-05-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Amino-Terminal Domain from the Tfb1 Subunit of TFIIH and
Characterization of Its Phosphoinositide and VP16 Binding Sites
Biochemistry, 44, 2005
|
|
2GS0
 
 | | NMR structure of the complex between the PH domain of the Tfb1 subunit from TFIIH and the activation domain of p53 | | Descriptor: | Cellular tumor antigen p53, RNA polymerase II transcription factor B subunit 1 | | Authors: | Di Lello, P, Jones, T.N, Nguyen, B.D, Legault, P, Omichinski, J.G. | | Deposit date: | 2006-04-25 | | Release date: | 2006-10-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the Tfb1/p53 complex: Insights into the interaction between the p62/Tfb1 subunit of TFIIH and the activation domain of p53.
Mol.Cell, 22, 2006
|
|
7K3Z
 
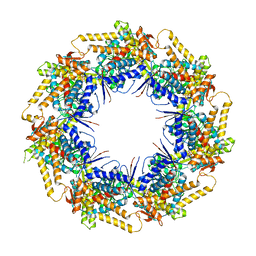 | | P. falciparum Cpn60 D474A mutant bound to ATP | | Descriptor: | 60 kDa chaperonin, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Tolia, N.H, Shi, D, Nguyen, B. | | Deposit date: | 2020-09-14 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.69 Å) | | Cite: | Crystal structure of P. falciparum Cpn60 bound to ATP reveals an open dynamic conformation before substrate binding.
Sci Rep, 11, 2021
|
|
