2W12
 
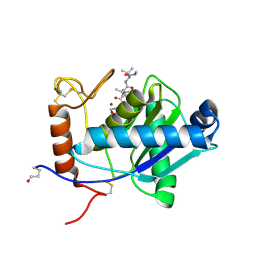 | | High-resolution crystal structure of the P-I snake venom metalloproteinase BaP1 in complex with a peptidomimetic: insights into inhibitor binding | | Descriptor: | (2R,3R)-N^1^-[(1S)-2,2-DIMETHYL-1-(METHYLCARBAMOYL)PROPYL]-N^4^-HYDROXY-2-(2-METHYLPROPYL)-3-{[(1,3-THIAZOL-2-YLCARBONYL)AMINO]METHYL}BUTANEDIAMIDE, GLYCEROL, ZINC ION, ... | | Authors: | Lingott, T.J, Schleberger, C, Gutierrez, J.M, Merfort, I. | | Deposit date: | 2008-10-14 | | Release date: | 2009-06-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | High-Resolution Crystal Structure of the Snake Venom Metalloproteinase Bap1 Complexed with a Peptidomimetic: Insight Into Inhibitor Binding.
Biochemistry, 48, 2009
|
|
2W14
 
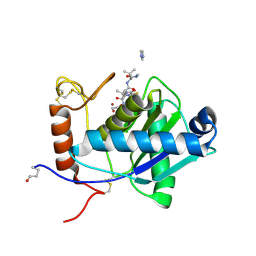 | | High-resolution crystal structure of the P-I snake venom metalloproteinase BaP1 in complex with a peptidomimetic: insights into inhibitor binding | | Descriptor: | (2R,3R)-N^1^-[(1S)-2,2-DIMETHYL-1-(METHYLCARBAMOYL)PROPYL]-N^4^-HYDROXY-2-(2-METHYLPROPYL)-3-{[(1,3-THIAZOL-2-YLCARBONYL)AMINO]METHYL}BUTANEDIAMIDE, GLYCEROL, IMIDAZOLE, ... | | Authors: | Lingott, T.J, Schleberger, C, Gutierrez, J.M, Merfort, I. | | Deposit date: | 2008-10-14 | | Release date: | 2009-06-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | High-Resolution Crystal Structure of the Snake Venom Metalloproteinase Bap1 Complexed with a Peptidomimetic: Insight Into Inhibitor Binding.
Biochemistry, 48, 2009
|
|
2W15
 
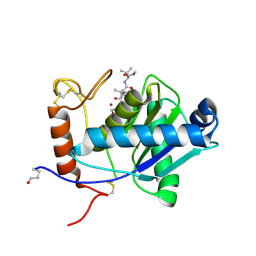 | | High-resolution crystal structure of the P-I snake venom metalloproteinase BaP1 in complex with a peptidomimetic: insights into inhibitor binding | | Descriptor: | (2R,3R)-N^1^-[(1S)-2,2-DIMETHYL-1-(METHYLCARBAMOYL)PROPYL]-N^4^-HYDROXY-2-(2-METHYLPROPYL)-3-{[(1,3-THIAZOL-2-YLCARBONYL)AMINO]METHYL}BUTANEDIAMIDE, GLYCEROL, ZINC ION, ... | | Authors: | Lingott, T.J, Schleberger, C, Gutierrez, J.M, Merfort, I. | | Deposit date: | 2008-10-14 | | Release date: | 2009-06-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | High-Resolution Crystal Structure of the Snake Venom Metalloproteinase Bap1 Complexed with a Peptidomimetic: Insight Into Inhibitor Binding.
Biochemistry, 48, 2009
|
|
2DW4
 
 | |
2DB3
 
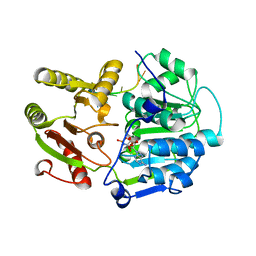 | | Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa | | Descriptor: | 5'-R(*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3', ATP-dependent RNA helicase vasa, MAGNESIUM ION, ... | | Authors: | Sengoku, T, Nureki, O, Nakamura, A, Kobayashi, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-14 | | Release date: | 2006-05-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa.
Cell(Cambridge,Mass.), 125, 2006
|
|
3AVS
 
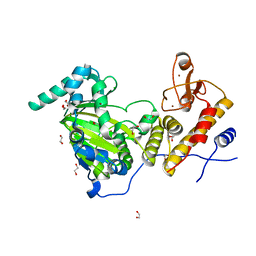 | | Catalytic fragment of UTX/KDM6A bound with N-oxyalylglycine, and Ni(II) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysine-specific demethylase 6A, ... | | Authors: | Sengoku, T, Yokoyama, S. | | Deposit date: | 2011-03-07 | | Release date: | 2011-10-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for histone H3 Lys 27 demethylation by UTX/KDM6A
Genes Dev., 25, 2011
|
|
3AVR
 
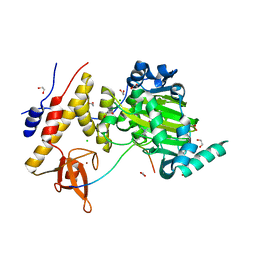 | | Catalytic fragment of UTX/KDM6A bound with histone H3K27me3 peptide, N-oxyalylglycine, and Ni(II) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Histone H3, ... | | Authors: | Sengoku, T, Yokoyama, S. | | Deposit date: | 2011-03-07 | | Release date: | 2011-10-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Structural basis for histone H3 Lys 27 demethylation by UTX/KDM6A
Genes Dev., 25, 2011
|
|
4TX1
 
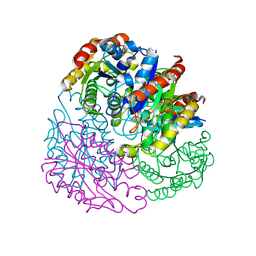 | |
3V67
 
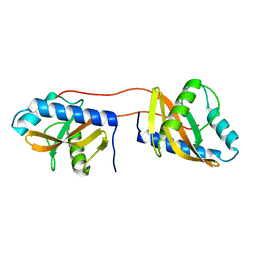 | | Periplasmic domain of Vibrio parahaemolyticus CpxA | | Descriptor: | Sensor protein CpxA | | Authors: | Kwon, E, Kim, D.Y, Ngo, T.D, Gross, J.D, Kim, K.K. | | Deposit date: | 2011-12-19 | | Release date: | 2012-09-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of the periplasmic domain of Vibrio parahaemolyticus CpxA
Protein Sci., 21, 2012
|
|
6MHE
 
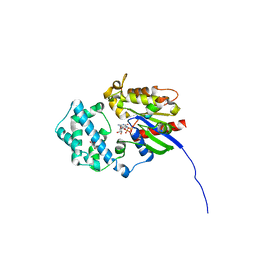 | | Galphai3 co-crystallized with KB752 | | Descriptor: | GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(k) subunit alpha, ... | | Authors: | Rees, S.D, Kalogriopoulos, N.A, Ngo, T, Kopcho, N, Ilatovskiy, A, Sun, N, Komives, E, Chang, G, Ghosh, P, Kufareva, I. | | Deposit date: | 2018-09-17 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for GPCR-independent activation of heterotrimeric Gi proteins.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6MHF
 
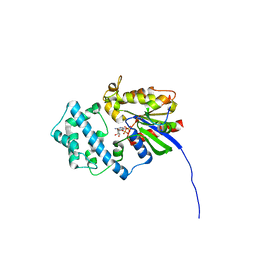 | | Galphai3 co-crystallized with GIV/Girdin | | Descriptor: | GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, Girdin, ... | | Authors: | Rees, S.D, Kalogriopoulos, N.A, Ngo, T, Kopcho, N, Ilatovskiy, A, Sun, N, Komives, E, Chang, G, Ghosh, P, Kufareva, I. | | Deposit date: | 2018-09-17 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for GPCR-independent activation of heterotrimeric Gi proteins.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4LAJ
 
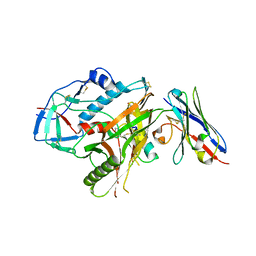 | | Crystal structure of HIV-1 YU2 envelope gp120 glycoprotein in complex with CD4-mimetic miniprotein, M48U1, and llama single-domain, broadly neutralizing, co-receptor binding site antibody, JM4 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-AMINOHEXANOIC ACID, ... | | Authors: | Acharya, P, Luongo, T.S, Kwong, P.D. | | Deposit date: | 2013-06-20 | | Release date: | 2013-08-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Heavy Chain-Only IgG2b Llama Antibody Effects Near-Pan HIV-1 Neutralization by Recognizing a CD4-Induced Epitope That Includes Elements of Coreceptor- and CD4-Binding Sites.
J.Virol., 87, 2013
|
|
8GUJ
 
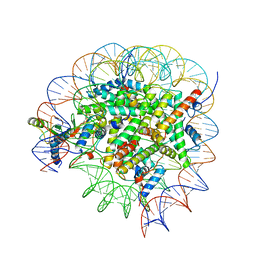 | | Bre1-nucleosome complex (Model II) | | Descriptor: | DNA (147-mer), E3 ubiquitin-protein ligase BRE1A, E3 ubiquitin-protein ligase BRE1B, ... | | Authors: | Onishi, S, Sato, K, Hamada, K, Nishizawa, T, Nureki, O, Ogata, K, Sengoku, T. | | Deposit date: | 2022-09-12 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of the human Bre1 complex bound to the nucleosome.
Nat Commun, 15, 2024
|
|
8GUK
 
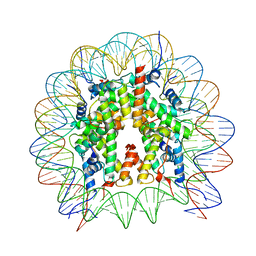 | | Human nucleosome core particle (free form) | | Descriptor: | DNA (147-mer), Histone H2A type 1, Histone H2B type 1-J, ... | | Authors: | Onishi, S, Sato, K, Nishizawa, T, Nureki, O, Ogata, K, Sengoku, T. | | Deposit date: | 2022-09-12 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Structure of the human Bre1 complex bound to the nucleosome.
Nat Commun, 15, 2024
|
|
8GUI
 
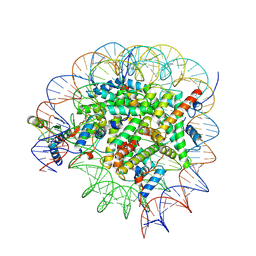 | | Bre1-nucleosome complex (Model I) | | Descriptor: | DNA (147-mer), E3 ubiquitin-protein ligase BRE1A, E3 ubiquitin-protein ligase BRE1B, ... | | Authors: | Onishi, S, Hamada, K, Sato, K, Nishizawa, T, Nureki, O, Ogata, K, Sengoku, T. | | Deposit date: | 2022-09-12 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structure of the human Bre1 complex bound to the nucleosome.
Nat Commun, 15, 2024
|
|
4RWY
 
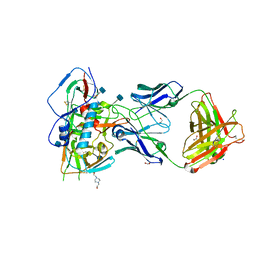 | | Crystal structure of VH1-46 germline-derived CD4-binding site-directed antibody 8ANC131 in complex with HIV-1 clade B YU2 gp120 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Acharya, P, Luongo, T.S, Kwong, P.D. | | Deposit date: | 2014-12-08 | | Release date: | 2015-07-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.128 Å) | | Cite: | Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors.
Cell(Cambridge,Mass.), 161, 2015
|
|
2E43
 
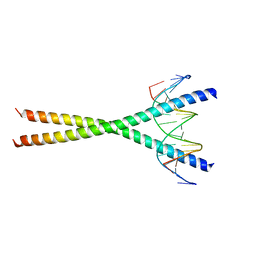 | | Crystal structure of C/EBPbeta Bzip homodimer K269A mutant bound to A High Affinity DNA fragment | | Descriptor: | CCAAT/enhancer-binding protein beta, DNA (5'-D(P*DAP*DAP*DTP*DAP*DTP*DTP*DGP*DCP*DGP*DCP*DAP*DAP*DTP*DCP*DCP*DT)-3'), DNA (5'-D(P*DTP*DAP*DGP*DGP*DAP*DTP*DTP*DGP*DCP*DGP*DCP*DAP*DAP*DTP*DAP*DT)-3') | | Authors: | Tahirov, T.H, Inoue-Bungo, T, Sato, K, Shiina, M, Hamada, K, Ogata, K. | | Deposit date: | 2006-12-01 | | Release date: | 2007-12-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Flexible Base Recognition by C/Ebpbeta
To be Published
|
|
2E42
 
 | | Crystal structure of C/EBPbeta Bzip homodimer V285A mutant bound to A High Affinity DNA fragment | | Descriptor: | CCAAT/enhancer-binding protein beta, DNA (5'-D(P*DAP*DAP*DTP*DAP*DTP*DTP*DGP*DCP*DGP*DCP*DAP*DAP*DTP*DCP*DCP*DT)-3'), DNA (5'-D(P*DTP*DAP*DGP*DGP*DAP*DTP*DTP*DGP*DCP*DGP*DCP*DAP*DAP*DTP*DAP*DT)-3') | | Authors: | Tahirov, T.H, Inoue-Bungo, T, Sato, K, Shiina, M, Hamada, K, Ogata, K. | | Deposit date: | 2006-12-01 | | Release date: | 2007-12-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Flexible Base Recognition by C/Ebpbeta
To be Published
|
|
8YE0
 
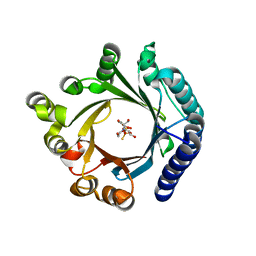 | | Crystal structure of KgpF prenyltransferase | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, LynF/TruF/PatF family peptide O-prenyltransferase, MAGNESIUM ION, ... | | Authors: | Hamada, K, Inoue, S, Goto, Y, Suga, H, Ogata, K, Sengoku, T. | | Deposit date: | 2024-02-21 | | Release date: | 2024-06-19 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | De Novo Discovery of Pseudo-Natural Prenylated Macrocyclic Peptide Ligands.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8JE4
 
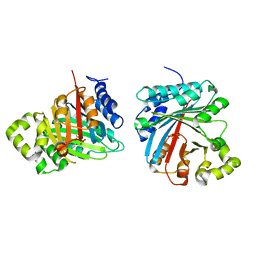 | | Crystal structure of LimF prenyltransferase (H239G/W273T mutant) bound with the thiodiphosphate moiety of farnesyl S-thiolodiphosphate (FSPP) | | Descriptor: | MAGNESIUM ION, TRIHYDROGEN THIODIPHOSPHATE, prenyltransferase, ... | | Authors: | Hamada, K, Oguni, A, Zhang, Y, Satake, M, Goto, Y, Suga, H, Ogata, K, Sengoku, T. | | Deposit date: | 2023-05-15 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Switching Prenyl Donor Specificities of Cyanobactin Prenyltransferases.
J.Am.Chem.Soc., 145, 2023
|
|
4JPW
 
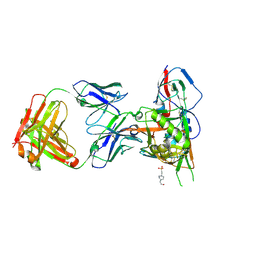 | | Crystal structure of broadly and potently neutralizing antibody 12a21 in complex with hiv-1 strain 93th057 gp120 mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEAVY CHAIN OF ANTIBODY 12A21, ... | | Authors: | Acharya, P, Luongo, T, Zhou, T, Kwong, P.D. | | Deposit date: | 2013-03-19 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.904 Å) | | Cite: | Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization.
Cell(Cambridge,Mass.), 153, 2013
|
|
8WM0
 
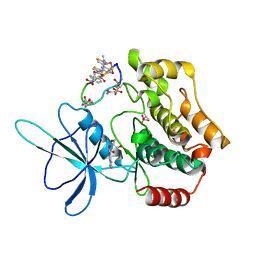 | | Crystal structure of TNIK-thiopeptide wTP3 complex | | Descriptor: | ADENOSINE, THIOPEPTIDE wTP3, TRAF2 and NCK-interacting protein kinase | | Authors: | Hamada, K, Kobayashi, S, Vinogradov, A.A, Zhang, Y, Goto, Y, Suga, H, Ogata, K, Sengoku, T. | | Deposit date: | 2023-10-01 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Compact Reprogrammed Genetic Code for De Novo Discovery of Proteolytically Stable Thiopeptides.
J.Am.Chem.Soc., 2024
|
|
2Z3Y
 
 | | Crystal structure of Lysine-specific demethylase1 | | Descriptor: | Lysine-specific histone demethylase 1, [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL (2R,3S,4S)-5-[7,8-DIMETHYL-2,4-DIOXO-5-(3-PHENYLPROPANOYL)-1,3,4,5-TETRAHYDROBENZO[G]PTERIDIN-10(2H)-YL]-2,3,4-TRIHYDROXYPENTYL DIHYDROGEN DIPHOSPHATE | | Authors: | Mimasu, S, Sengoku, T, Umehara, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-06-08 | | Release date: | 2008-01-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of histone demethylase LSD1 and tranylcypromine at 2.25A
Biochem.Biophys.Res.Commun., 366, 2008
|
|
3AXS
 
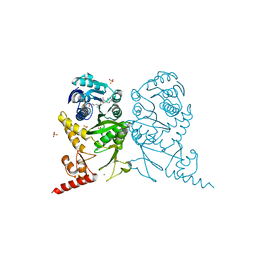 | | Complex structure of tRNA methyltransferase Trm1 from Aquifex aeolicus with sinefungin | | Descriptor: | Probable N(2),N(2)-dimethylguanosine tRNA methyltransferase Trm1, SINEFUNGIN, SULFATE ION, ... | | Authors: | Ihsanawati, Sengoku, T, Yokoyama, S, Bessho, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-04-13 | | Release date: | 2011-08-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.162 Å) | | Cite: | Substrate tRNA recognition mechanism of a multisite-specific tRNA methyltransferase, Aquifex aeolicus Trm1, based on the X-ray crystal structure
J.Biol.Chem., 286, 2011
|
|
3AXT
 
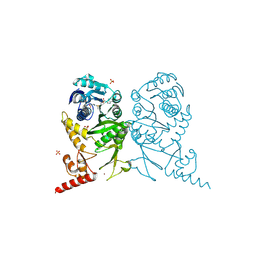 | | Complex structure of tRNA methyltransferase Trm1 from Aquifex aeolicus with S-adenosyl-L-Methionine | | Descriptor: | Probable N(2),N(2)-dimethylguanosine tRNA methyltransferase Trm1, S-ADENOSYLMETHIONINE, SULFATE ION, ... | | Authors: | Ihsanawati, Sengoku, T, Yokoyama, S, Bessho, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-04-14 | | Release date: | 2011-08-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.491 Å) | | Cite: | Substrate tRNA recognition mechanism of a multisite-specific tRNA methyltransferase, Aquifex aeolicus Trm1, based on the X-ray crystal structure
J.Biol.Chem., 286, 2011
|
|
