4NOA
 
 | |
4NPH
 
 | | Crystal structure of SsaN from Salmonella enterica | | Descriptor: | Probable secretion system apparatus ATP synthase SsaN | | Authors: | Sugiman-Marangos, S.N, Zhang, K, Cooper, C.A, Coombes, B.K, Junop, M.S. | | Deposit date: | 2013-11-21 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of SsaN from Salmonella enterica
J.Biol.Chem., 2014
|
|
1K1G
 
 | | STRUCTURAL BASIS FOR RECOGNITION OF THE INTRON BRANCH SITE RNA BY SPLICING FACTOR 1 | | Descriptor: | 5'-R(*UP*AP*UP*AP*CP*UP*AP*AP*CP*AP*A)-3', SF1-Bo isoform | | Authors: | Liu, Z, Luyten, I, Bottomley, M.J, Messias, A.C, Houngninou-Molango, S, Sprangers, R, Zanier, K, Kramer, A, Sattler, M. | | Deposit date: | 2001-09-25 | | Release date: | 2001-11-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recognition of the intron branch site RNA by splicing factor 1.
Science, 294, 2001
|
|
4MBQ
 
 | | TPR3 of FimV from P. aeruginosa (PAO1) | | Descriptor: | Motility protein FimV | | Authors: | Nguyen, Y, Zhang, K, Daniel-Ivad, M, Robinson, H, Wolfram, F, Sugiman-Marangos, S.N, Junop, M.S, Burrows, L.L, Howell, P.L. | | Deposit date: | 2013-08-19 | | Release date: | 2014-08-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Crystal structure of TPR2 from FimV
To be Published
|
|
4NOE
 
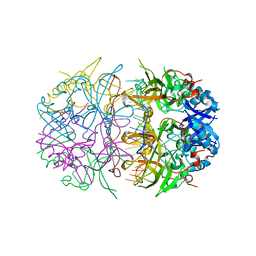 | | Crystal structure of DdrB bound to 30b ssDNA | | Descriptor: | 5'-D(*TP*TP*GP*CP*GP*CP*TP*TP*GP*CP*GP*CP*TP*TP*GP*CP*GP*CP*TP*TP*GP*CP*GP*CP*TP*TP*GP*CP*GP*CP)-3', CALCIUM ION, Single-stranded DNA-binding protein DdrB | | Authors: | Sugiman-Marangos, S.N, Weiss, Y.M, Junop, M.S. | | Deposit date: | 2013-11-19 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism for accurate, protein-assisted DNA annealing by Deinococcus radiodurans DdrB.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5GIK
 
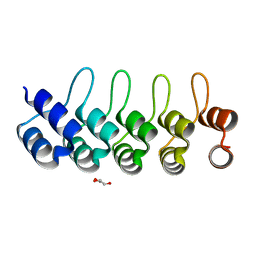 | | Modulation of the affinity of a HIV-1 capsid-directed ankyrin towards its viral target through critical amino acid editing | | Descriptor: | Artificial ankyrin repeat protein_Ank(GAG)1D4 mutant -S45Y, GLYCEROL | | Authors: | Chuankhayan, P, Saoin, S, Wisitponchai, T, Intachai, K, Chupradit, K, Moonmuang, S, Kitidee, K, Nangola, S, Sanghiran, L.V, Hong, S.S, Boulanger, P, Tayapiwatana, C, Chen, C.J. | | Deposit date: | 2016-06-24 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Modulation of the affinity of a HIV-1 capsid-directed ankyrintowards its viral target through critical amino acid editing
To Be Published
|
|
6KGK
 
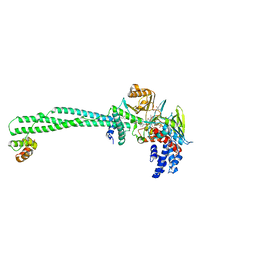 | | LSD1-CoREST-S2101 five-membered ring adduct model | | Descriptor: | 3-[3,5-bis(fluoranyl)-2-phenylmethoxy-phenyl]propanal, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Niwa, H, Sato, S, Sengoku, S, Umehara, T, Yokoyama, S. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Development and Structural Evaluation of N-Alkylated trans-2-Phenylcyclopropylamine-Based LSD1 Inhibitors.
Chemmedchem, 15, 2020
|
|
4MAL
 
 | | TPR3 of FimV from P. aeruginosa (PAO1) | | Descriptor: | Motility protein FimV | | Authors: | Nguyen, Y, Zhang, K, Daniel-Ivad, M, Sugiman-Marangos, S.N, Junop, M.S, Burrows, L.L, Howell, P.L. | | Deposit date: | 2013-08-16 | | Release date: | 2014-08-20 | | Last modified: | 2016-02-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of TPR2 from FimV
To be Published
|
|
4L8M
 
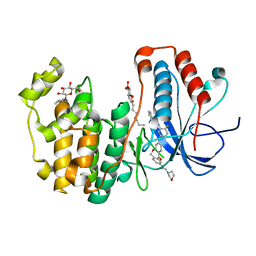 | | Human p38 MAP kinase in complex with a Dibenzoxepinone | | Descriptor: | Mitogen-activated protein kinase 14, N-[2-fluoro-5-({9-[2-(morpholin-4-yl)ethoxy]-11-oxo-6,11-dihydrodibenzo[b,e]oxepin-3-yl}amino)phenyl]benzamide, octyl beta-D-glucopyranoside | | Authors: | Richters, A, Mayer-Wrangowski, S.C, Gruetter, C, Rauh, D. | | Deposit date: | 2013-06-17 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Metabolically Stable Dibenzo[b,e]oxepin-11(6H)-ones as Highly Selective p38 MAP Kinase Inhibitors: Optimizing Anti-Cytokine Activity in Human Whole Blood.
J.Med.Chem., 56, 2013
|
|
3UVR
 
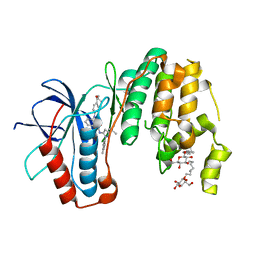 | | Human p38 MAP Kinase in Complex with KM064 | | Descriptor: | 1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]-3-{3-[(5-oxo-6,7,8,9-tetrahydro-5H-benzo[7]annulen-2-yl)amino]phenyl}urea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Richters, A, Mayer-Wrangowski, S.C, Gruetter, C, Rauh, D. | | Deposit date: | 2011-11-30 | | Release date: | 2012-12-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: |
|
|
1QFD
 
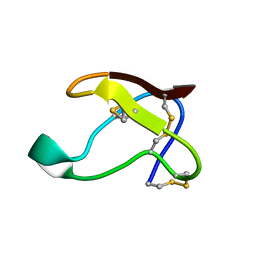 | | NMR SOLUTION STRUCTURE OF ALPHA-AMYLASE INHIBITOR (AAI) | | Descriptor: | PROTEIN (ALPHA-AMYLASE INHIBITOR) | | Authors: | Lu, S, Deng, P, Liu, X, Luo, J, Han, R, Gu, X, Liang, S, Wang, X, Feng, L, Lozanov, V, Patthy, A, Pongor, S. | | Deposit date: | 1999-04-08 | | Release date: | 1999-07-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the major alpha-amylase inhibitor of the crop plant amaranth.
J.Biol.Chem., 274, 1999
|
|
1NPI
 
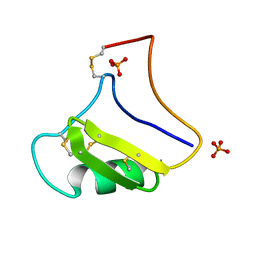 | | Tityus Serrulatus Neurotoxin (Ts1) at atomic resolution | | Descriptor: | PHOSPHATE ION, Toxin VII | | Authors: | Pinheiro, C.B, Marangoni, S, Toyama, M.H, Polikarpov, I. | | Deposit date: | 2003-01-17 | | Release date: | 2003-02-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structural analysis of Tityus serrulatus Ts1 neurotoxin at atomic resolution: insights into interactions with Na+ channels.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1CLV
 
 | | YELLOW MEAL WORM ALPHA-AMYLASE IN COMPLEX WITH THE AMARANTH ALPHA-AMYLASE INHIBITOR | | Descriptor: | CALCIUM ION, CHLORIDE ION, PROTEIN (ALPHA-AMYLASE INHIBITOR), ... | | Authors: | Pereira, P.J.B, Lozanov, V, Patthy, A, Huber, R, Bode, W, Pongor, S, Strobl, S. | | Deposit date: | 1999-05-04 | | Release date: | 2000-05-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specific inhibition of insect alpha-amylases: yellow meal worm alpha-amylase in complex with the amaranth alpha-amylase inhibitor at 2.0 A resolution.
Structure Fold.Des., 7, 1999
|
|
4HLL
 
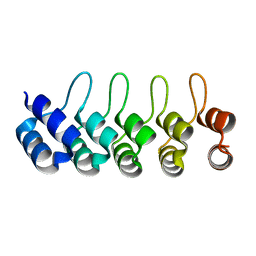 | | Crystal structure of Artificial ankyrin repeat protein_Ank(GAG)1D4 | | Descriptor: | Ankyrin(GAG)1D4 | | Authors: | Chuankhayan, P, Nangola, S, Minard, P, Boulanger, P, Hong, S.S, Tayapiwatana, C, Chen, C.-J. | | Deposit date: | 2012-10-17 | | Release date: | 2013-10-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of Gag bioactive determinants specific to designed ankyrin and interfering in HIV-1 assembly
To be Published
|
|
1R8N
 
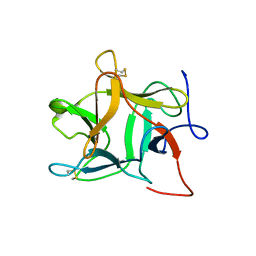 | |
1R8O
 
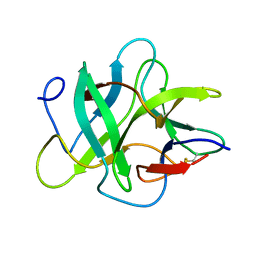 | | Crystal structure of an unusual Kunitz-type trypsin inhibitor from Copaifera langsdorffii seeds | | Descriptor: | Kunitz trypsin inhibitor | | Authors: | Krauchenco, S, Nagem, R.A.P, da Silva, J.A, Marangoni, S, Polikarpov, I. | | Deposit date: | 2003-10-27 | | Release date: | 2004-05-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Three-dimensional structure of an unusual Kunitz (STI) type trypsin inhibitor from Copaifera langsdorffii.
Biochimie, 86, 2004
|
|
2KB9
 
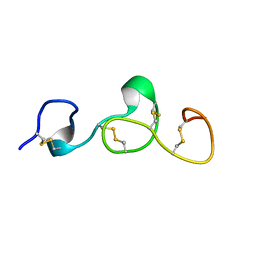 | |
2MH3
 
 | | NMR structure of the basic helix-loop-helix region of the transcriptional repressor HES-1 | | Descriptor: | Transcription factor HES-1 | | Authors: | Wienk, H, Popovic, M, Coglievina, M, Boelens, R, Pongor, S, Pintar, A. | | Deposit date: | 2013-11-15 | | Release date: | 2014-02-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The basic helix-loop-helix region of the transcriptional repressor hairy and enhancer of split 1 is preorganized to bind DNA.
Proteins, 82, 2014
|
|
