5G26
 
 | | Unveiling the Mechanism Behind the in-meso Crystallization of Membrane Proteins | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-hexadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-hexadec-9-enoate, INTIMIN | | 著者 | Zabara, A, Newman, J, Peat, T.S. | | 登録日 | 2016-04-07 | | 公開日 | 2017-02-15 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.42 Å) | | 主引用文献 | The Nanoscience Behind the Art of in-Meso Crystallization of Membrane Proteins.
Nanoscale, 9, 2017
|
|
5HY4
 
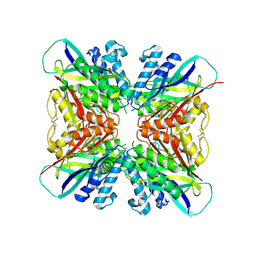 | | Structure-function analysis of functionally diverse members of the cyclic amide hydrolase family of Toblerone fold enzymes | | 分子名称: | Ring-opening amidohydrolase | | 著者 | Peat, T.S, Balotra, S, Wilding, M, Newman, J, Scott, C. | | 登録日 | 2016-02-01 | | 公開日 | 2017-02-01 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | High-Resolution X-Ray Structures of Two Functionally Distinct Members of the Cyclic Amide Hydrolase Family of Toblerone Fold Enzymes.
Appl. Environ. Microbiol., 83, 2017
|
|
5HY0
 
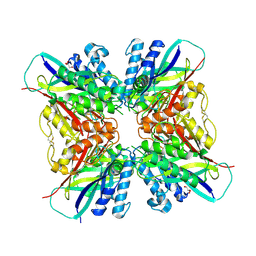 | | orotic acid hydrolase | | 分子名称: | GLYCEROL, Ring-opening amidohydrolase | | 著者 | Peat, T.S, Balotra, S, Wilding, M, Newman, J, Scott, C. | | 登録日 | 2016-02-01 | | 公開日 | 2017-02-01 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | High-Resolution X-Ray Structures of Two Functionally Distinct Members of the Cyclic Amide Hydrolase Family of Toblerone Fold Enzymes.
Appl. Environ. Microbiol., 83, 2017
|
|
7PSP
 
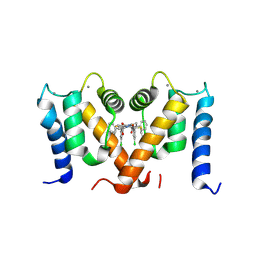 | | Crystal structure of S100A4 labeled with NU000846b. | | 分子名称: | (2R,4R)-1-(2-chloranylethanoyl)-N-(3-chlorophenyl)-4-phenyl-pyrrolidine-2-carboxamide, CALCIUM ION, Protein S100-A4 | | 著者 | Giroud, C, Szommer, T, Coxon, C, Monteiro, O, Christott, T, Bennett, J, Aitmakhanova, K, Raux, B, Newman, J, Elkins, J, Arruda Bezerra, G, Krojer, T, Koekemoer, L, Von Delft, F, Bountr, C, Brennan, P, Fedorov, O. | | 登録日 | 2021-09-23 | | 公開日 | 2022-10-05 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Crystal structure of S100A4 labeled with NU000846b.
To Be Published
|
|
3PN1
 
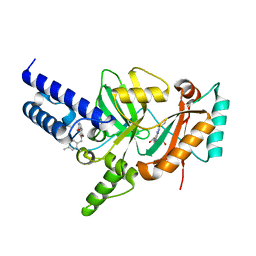 | | Novel Bacterial NAD+-dependent DNA Ligase Inhibitors with Broad Spectrum Potency and Antibacterial Efficacy In Vivo | | 分子名称: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 2-(butylsulfanyl)adenosine, DNA ligase | | 著者 | Mills, S, Eakin, A, Buurman, E, Newman, J, Gao, N, Huynh, H, Johnson, K, Lahiri, S, Shapiro, A, Walkup, G, Wei, Y, Stokes, S. | | 登録日 | 2010-11-18 | | 公開日 | 2011-01-12 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Novel Bacterial NAD+-Dependent DNA Ligase Inhibitors with Broad-Spectrum Activity and Antibacterial Efficacy In Vivo.
Antimicrob.Agents Chemother., 55, 2011
|
|
6G0Q
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with an acetylated GATA1 peptide (K312ac/K315ac) | | 分子名称: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, Erythroid transcription factor | | 著者 | Filippakopoulos, P, Picaud, S, Newman, J, Sorrell, F, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C. | | 登録日 | 2018-03-19 | | 公開日 | 2018-11-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol. Cell, 73, 2019
|
|
6G0O
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with an acetylated ATRX peptide (K1030ac/K1033ac) | | 分子名称: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, Transcriptional regulator ATRX | | 著者 | Filippakopoulos, P, Picaud, S, Newman, J, Sorrell, F, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C. | | 登録日 | 2018-03-19 | | 公開日 | 2018-11-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol. Cell, 73, 2019
|
|
5NNG
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with an acetylated SRPK1 peptide (K585ac) | | 分子名称: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, VAG(ALY)YS(ALY)EFFY | | 著者 | Filippakopoulos, P, Picaud, S, Newman, J, Sorrell, F, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Structural Genomics Consortium (SGC) | | 登録日 | 2017-04-08 | | 公開日 | 2018-05-16 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol. Cell, 73, 2019
|
|
5FDP
 
 | | Structure of DDR1 receptor tyrosine kinase in complex with D2099 inhibitor at 2.25 Angstroms resolution. | | 分子名称: | (4~{S})-4-methyl-~{N}-[3-[(4-methylpiperazin-1-yl)methyl]-5-(trifluoromethyl)phenyl]-2-pyrimidin-5-yl-3,4-dihydro-1~{H}-isoquinoline-7-carboxamide, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Bartual, S.G, Pinkas, D.M, Wang, Z, Ding, K, Mahajan, P, Kupinska, K, Mukhopadhyay, S, Strain-Damerell, C, Borkowska, O, Talon, R, Kopec, J, Williams, E, Tallant, C, Chaikuad, A, Sorell, F, Newman, J, Burgess-Brown, N, Arrowsmith, C.H, von Delft, F, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | 登録日 | 2015-12-16 | | 公開日 | 2016-06-08 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structure-Based Design of Tetrahydroisoquinoline-7-carboxamides as Selective Discoidin Domain Receptor 1 (DDR1) Inhibitors.
J.Med.Chem., 59, 2016
|
|
6FNU
 
 | | Structure of S. cerevisiae Methylenetetrahydrofolate reductase 1, catalytic domain | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Methylenetetrahydrofolate reductase 1 | | 著者 | Kopec, J, Rembeza, E, Bezerra, G.A, Newman, J, Bountra, C, Froese, D.S, Baumgartner, M, Yue, W.W, Structural Genomics Consortium (SGC) | | 登録日 | 2018-02-05 | | 公開日 | 2018-03-07 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Structural basis for the regulation of human 5,10-methylenetetrahydrofolate reductase by phosphorylation and S-adenosylmethionine inhibition.
Nat Commun, 9, 2018
|
|
5FDX
 
 | | Structure of DDR1 receptor tyrosine kinase in complex with D2164 inhibitor at 2.65 Angstroms resolution. | | 分子名称: | 1,2-ETHANEDIOL, 3-[(4-methylpiperazin-1-yl)methyl]-~{N}-[(4~{R})-4-methyl-2-pyrimidin-5-yl-3,4-dihydro-1~{H}-isoquinolin-7-yl]-5-(trifluoromethyl)benzamide, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Bartual, S.G, Pinkas, D.M, Wang, Z, Ding, K, Mahajan, P, Kupinska, K, Mukhopadhyay, S, Strain-Damerell, C, Chalk, R, Borkowska, O, Talon, R, Kopec, J, Williams, E, Tallant, C, Chaikuad, A, Sorell, F, Newman, J, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | 登録日 | 2015-12-16 | | 公開日 | 2016-10-26 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structure of DDR1 receptor tyrosine kinase in complex with D2164 inhibitor at 2.65 Angstroms resolution.
To Be Published
|
|
6G4Q
 
 | | Structure of human ADP-forming succinyl-CoA ligase complex SUCLG1-SUCLA2 | | 分子名称: | 1,2-ETHANEDIOL, Succinate--CoA ligase [ADP-forming] subunit beta, mitochondrial, ... | | 著者 | Bailey, H.J, Shrestha, L, Rembeza, E, Sorrell, F.J, Newman, J, Strain-Damerell, C, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W. | | 登録日 | 2018-03-28 | | 公開日 | 2018-04-11 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Structure of human ADP-forming succinyl-CoA ligase complex SUCLG1-SUCLA2
To Be Published
|
|
5LTU
 
 | | Crystal Structure of NUDT4A- Diphosphoinositol polyphosphate phosphohydrolase 2 | | 分子名称: | 1,2-ETHANEDIOL, Diphosphoinositol polyphosphate phosphohydrolase 2 | | 著者 | Srikannathasan, V, Nunez, C.A, Tallant, C, Siejka, P, Mathea, S, Newman, J, Strain-Damerell, C, Elkins, J.M, Burgess-Brown, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Huber, K. | | 登録日 | 2016-09-07 | | 公開日 | 2017-09-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | Crystal Structure of Human NUDT4A- Diphosphoinositol polyphosphate phosphohydrolase 2
To Be Published
|
|
5MQ4
 
 | | Crystal Structure of the leucine zipper of human PRKCBP1 | | 分子名称: | Protein kinase C-binding protein 1, SULFATE ION, ZINC ION | | 著者 | Krojer, T, Savitsky, P, Picaud, S, Newman, J, Tallant, C, Heroven, C, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Filippakopoulos, P. | | 登録日 | 2016-12-20 | | 公開日 | 2017-01-25 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal Structure of the leucine zipper of human PRKCBP1
To Be Published
|
|
4CQB
 
 | | The reaction mechanism of the N-isopropylammelide isopropylaminohydrolase AtzC: insights from structural and mutagenesis studies | | 分子名称: | CHLORIDE ION, DIMETHYL SULFOXIDE, MALONATE ION, ... | | 著者 | Balotra, S, Newman, J, French, N.G, Peat, T.S, Scott, C. | | 登録日 | 2014-02-13 | | 公開日 | 2015-03-04 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | X-Ray Structure and Mutagenesis Studies of the N-Isopropylammelide Isopropylaminohydrolase, Atzc
Plos One, 1, 2015
|
|
4CNR
 
 | | Surface residue engineering of bovine carbonic anhydrase to an extreme halophilic enzyme for potential application in postcombustion CO2 capture | | 分子名称: | CARBONIC ANHYDRASE 2, ZINC ION | | 著者 | Warden, A, Newman, J, Peat, T.S, Seabrook, S, Williams, M, Dojchinov, G, Haritos, V. | | 登録日 | 2014-01-24 | | 公開日 | 2015-02-04 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Rational Engineering of a Mesohalophilic Carbonic Anhydrase to an Extreme Halotolerant Biocatalyst.
Nat.Commun., 6, 2015
|
|
4CYY
 
 | | The structure of vanin-1: defining the link between metabolic disease, oxidative stress and inflammation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PANTETHEINASE | | 著者 | Boersma, Y.L, Newman, J, Adams, T.E, Sparrow, L, Cowieson, N, Lucent, D, Krippner, G, Bozaoglu, K, Peat, T.S. | | 登録日 | 2014-04-16 | | 公開日 | 2014-12-10 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | The Structure of Vanin-1: A Key Enzyme Linking Metabolic Disease and Inflammation
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4CYG
 
 | | The structure of vanin-1: defining the link between metabolic disease, oxidative stress and inflammation | | 分子名称: | (2R)-2,4-dihydroxy-N-[(3S)-3-hydroxy-4-phenylbutyl]-3,3-dimethylbutanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Boersma, Y.L, Newman, J, Adams, T.E, Sparrow, L, Cowieson, N, Lucent, D, Krippner, G, Bozaoglu, K, Peat, T.S. | | 登録日 | 2014-04-11 | | 公開日 | 2014-12-10 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The Structure of Vanin-1: A Key Enzyme Linking Metabolic Disease and Inflammation
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4CYF
 
 | | The structure of vanin-1: defining the link between metabolic disease, oxidative stress and inflammation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PANTETHEINASE | | 著者 | Boersma, Y.L, Newman, J, Adams, T.E, Sparrow, L, Cowieson, N, Lucent, D, Krippner, G, Bozaoglu, K, Peat, T.S. | | 登録日 | 2014-04-11 | | 公開日 | 2014-12-10 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | The Structure of Vanin-1: A Key Enzyme Linking Metabolic Disease and Inflammation
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4CNW
 
 | | Surface residue engineering of bovine carbonic anhydrase to an extreme halophilic enzyme for potential application in postcombustion CO2 capture | | 分子名称: | CALCIUM ION, CARBONIC ANHYDRASE 2, ZINC ION | | 著者 | Warden, A, Newman, J, Peat, T.S, Seabrook, S, Williams, M, Dojchinov, G, Haritos, V. | | 登録日 | 2014-01-25 | | 公開日 | 2015-02-04 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Rational Engineering of a Mesohalophilic Carbonic Anhydrase to an Extreme Halotolerant Biocatalyst.
Nat.Commun., 6, 2015
|
|
4BVS
 
 | | Cyanuric acid hydrolase: evolutionary innovation by structural concatenation. | | 分子名称: | 1,3,5-triazine-2,4,6-triamine, CYANURIC ACID AMIDOHYDROLASE, MAGNESIUM ION | | 著者 | Peat, T.S, Balotra, S, Wilding, M, French, N.G, Briggs, L.J, Panjikar, S, Cowieson, N, Newman, J, Scott, C. | | 登録日 | 2013-06-28 | | 公開日 | 2013-07-17 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Cyanuric Acid Hydrolase: Evolutionary Innovation by Structural Concatenation.
Mol.Microbiol., 88, 2013
|
|
4BVT
 
 | | Cyanuric acid hydrolase: evolutionary innovation by structural concatenation | | 分子名称: | BARBITURIC ACID, CYANURIC ACID AMIDOHYDROLASE, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Peat, T.S, Balotra, S, Wilding, M, French, N.G, Briggs, L.J, Panjikar, S, Cowieson, N, Newman, J, Scott, C. | | 登録日 | 2013-06-28 | | 公開日 | 2013-07-17 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Cyanuric Acid Hydrolase: Evolutionary Innovation by Structural Concatenation.
Mol.Microbiol., 88, 2013
|
|
4CP8
 
 | | Structure of the amidase domain of allophanate hydrolase from Pseudomonas sp strain ADP | | 分子名称: | ALLOPHANATE HYDROLASE, MALONATE ION | | 著者 | Balotra, S, Newman, J, French, N, French, L, Peat, T.S, Scott, C. | | 登録日 | 2014-02-03 | | 公開日 | 2014-11-26 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | X-Ray Structure of the Amidase Domain of Atzf, the Allophanate Hydrolase from the Cyanuric Acid-Mineralizing Multienzyme Complex.
Appl.Environ.Microbiol., 81, 2015
|
|
4CQD
 
 | | The reaction mechanism of the N-isopropylammelide isopropylaminohydrolase AtzC: insights from structural and mutagenesis studies | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, MALONATE ION, ... | | 著者 | Balotra, S, Newman, J, French, N.G, Peat, T.S, Scott, C. | | 登録日 | 2014-02-13 | | 公開日 | 2015-03-04 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | X-Ray Structure and Mutagenesis Studies of the N-Isopropylammelide Isopropylaminohydrolase, Atzc
Plos One, 1, 2015
|
|
5TI8
 
 | |
