5T46
 
 | | Crystal structure of the human eIF4E-eIF4G complex | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, Eukaryotic translation initiation factor 4 gamma 1, Eukaryotic translation initiation factor 4E, ... | | Authors: | Gruener, S, Peter, D, Weber, R, Wohlbold, L, Chung, M.-Y, Weichenrieder, O, Valkov, E, Igreja, C, Izaurralde, E. | | Deposit date: | 2016-08-29 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | The Structures of eIF4E-eIF4G Complexes Reveal an Extended Interface to Regulate Translation Initiation.
Mol.Cell, 64, 2016
|
|
4BMW
 
 | | Crystal structure of the Streptomyces reticuli HbpS E78D, E81D double mutant | | Descriptor: | EXTRACELLULAR HAEM-BINDING PROTEIN | | Authors: | Wagener, S, Kursula, I, Wedderhoff, I, Groves, M.R, Ortiz de Orue Lucana, D. | | Deposit date: | 2013-05-11 | | Release date: | 2013-09-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Iron Binding at Specific Sites within the Octameric Hbps Protects Streptomycetes from Iron-Mediated Oxidative Stress.
Plos One, 8, 2013
|
|
5T48
 
 | | Crystal structure of the D. melanogaster eIF4E-eIF4G complex without lateral contact | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, Eukaryotic translation initiation factor 4E, Eukaryotic translation initiation factor 4G, ... | | Authors: | Gruener, S, Peter, D, Weber, R, Wohlbold, L, Chung, M.-Y, Weichenrieder, O, Valkov, E, Igreja, C, Izaurralde, E. | | Deposit date: | 2016-08-29 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The Structures of eIF4E-eIF4G Complexes Reveal an Extended Interface to Regulate Translation Initiation.
Mol.Cell, 64, 2016
|
|
4NSL
 
 | |
5E0F
 
 | | Human pancreatic alpha-amylase in complex with mini-montbretin A | | Descriptor: | 5,7-dihydroxy-4-oxo-2-(3,4,5-trihydroxyphenyl)-4H-chromen-3-yl 6-deoxy-2-O-{6-O-[(2E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]-beta-D-glucopyranosyl}-alpha-L-mannopyranoside, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Caner, S, Brayer, G.D. | | Deposit date: | 2015-09-28 | | Release date: | 2016-09-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Human pancreatic alpha-amylase in complex with mini-montbretin A
To Be Published
|
|
5U3A
 
 | |
5VA9
 
 | |
5TD4
 
 | |
6OCN
 
 | |
6OBX
 
 | |
4JUZ
 
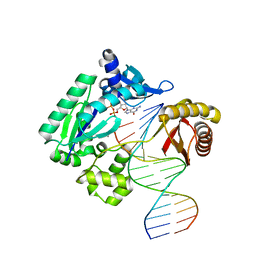 | | Ternary complex of gamma-OHPDG adduct modified dna (zero primer) with dna polymerase iv and incoming dgtp | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*CP*C)-3'), ... | | Authors: | Banerjee, S, Shanmugam, G, Stone, M.P. | | Deposit date: | 2013-03-25 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Ring-Opening of the gamma-OH-PdG Adduct Promotes Error-Free Bypass by the Sulfolobus solfataricus DNA Polymerase Dpo4.
Chem.Res.Toxicol., 26, 2013
|
|
4JV2
 
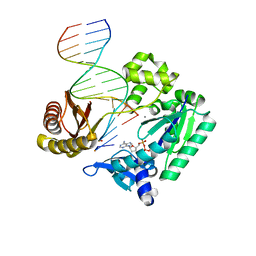 | | Ternary complex of gamma-OHPDG adduct modified dna with dna (-1 primer) polymerase iv and incoming datp | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*C)-3'), ... | | Authors: | Banerjee, S, Shanmugam, G, Stone, M.P. | | Deposit date: | 2013-03-25 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Ring-Opening of the gamma-OH-PdG Adduct Promotes Error-Free Bypass by the Sulfolobus solfataricus DNA Polymerase Dpo4.
Chem.Res.Toxicol., 26, 2013
|
|
4JV0
 
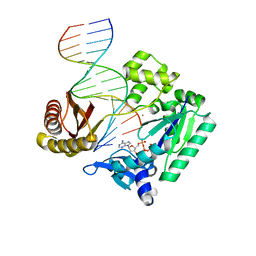 | | Ring-Opening of the -OH-PdG Adduct in Ternary Complexes with the Sulfolobus solfataricus DNA polymerase Dpo4 | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*CP*C)-3'), ... | | Authors: | Banerjee, S, Shanmugam, G, Stone, M.P. | | Deposit date: | 2013-03-25 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Ring-Opening of the gamma-OH-PdG Adduct Promotes Error-Free Bypass by the Sulfolobus solfataricus DNA Polymerase Dpo4.
Chem.Res.Toxicol., 26, 2013
|
|
4JV1
 
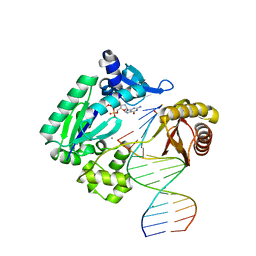 | | Ternary complex of gamma-OHPDG adduct modified dna with dna (-1 primer) polymerase iv and incoming dgtp | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*C)-3'), ... | | Authors: | Banerjee, S, Shanmugam, G, Stone, M.P. | | Deposit date: | 2013-03-25 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ring-Opening of the gamma-OH-PdG Adduct Promotes Error-Free Bypass by the Sulfolobus solfataricus DNA Polymerase Dpo4.
Chem.Res.Toxicol., 26, 2013
|
|
1ZP2
 
 | |
8QCL
 
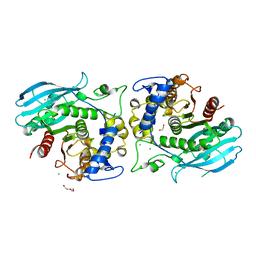 | | A carbohydrate esterase family 15 (CE15) glucuronoyl esterase from Phocaeicola vulgatus ATCC 8482 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Banerjee, S, Poulsen, J.N, Mazurkewich, S, Seveso, A, Larsbrink, J, Lo Leggio, L. | | Deposit date: | 2023-08-27 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Polysaccharide utilization loci from Bacteroidota encode CE15 enzymes with possible roles in cleaving pectin-lignin bonds.
Appl.Environ.Microbiol., 90, 2024
|
|
8QEF
 
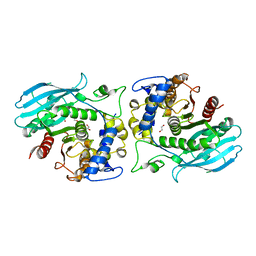 | | A carbohydrate esterase family 15 (CE15) glucuronoyl esterase from Phocaeicola ATCC 8482 bound to novel ligand. | | Descriptor: | 1,2-ETHANEDIOL, Putative acetyl xylan esterase, beta-D-galactopyranuronic acid | | Authors: | Banerjee, S, Poulsen, J.N, Mazurkewich, S, Seveso, A, Larsbrink, J, Lo Leggio, L. | | Deposit date: | 2023-08-31 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Polysaccharide utilization loci from Bacteroidota encode CE15 enzymes with possible roles in cleaving pectin-lignin bonds.
Appl.Environ.Microbiol., 90, 2024
|
|
1O6W
 
 | |
8QY4
 
 | | Structure of interleukin 11 (gp130 P496L mutant). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-11, ... | | Authors: | Gardner, S, Bubeck, D, Jin, Y. | | Deposit date: | 2023-10-25 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structure of interleukin 11 (gp130 P496L mutant).
To Be Published
|
|
8QY5
 
 | | Structure of interleukin 6. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-6, ... | | Authors: | Gardner, S, Bubeck, D, Jin, Y. | | Deposit date: | 2023-10-25 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of interleukin 6.
To Be Published
|
|
8QY6
 
 | | Structure of interleukin 6 (gp130 P496L mutant). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-6, ... | | Authors: | Gardner, S, Bubeck, D, Jin, Y. | | Deposit date: | 2023-10-25 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structure of interleukin 6 (gp130 P496L mutant).
To Be Published
|
|
1NR1
 
 | | Crystal structure of the R463A mutant of human Glutamate dehydrogenase | | Descriptor: | Glutamate dehydrogenase 1 | | Authors: | Banerjee, S, Schmidt, T, Fang, J, Stanley, C.A, Smith, T.J. | | Deposit date: | 2003-01-23 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural studies on ADP activation of mammalian glutamate dehydrogenase and the evolution of regulation
Biochemistry, 42, 2003
|
|
1NQT
 
 | | Crystal structure of bovine Glutamate dehydrogenase-ADP complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glutamate dehydrogenase 1 | | Authors: | Banerjee, S, Schmidt, T, Fang, J, Stanley, C.A, Smith, T.J. | | Deposit date: | 2003-01-23 | | Release date: | 2003-05-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural studies on ADP activation of mammalian glutamate dehydrogenase and the evolution of regulation
Biochemistry, 42, 2003
|
|
1OME
 
 | | CRYSTAL STRUCTURE OF THE OMEGA LOOP DELETION MUTANT (RESIDUES 163-178 DELETED) OF BETA-LACTAMASE FROM STAPHYLOCOCCUS AUREUS PC1 | | Descriptor: | BETA-LACTAMASE, CHLORIDE ION | | Authors: | Banerjee, S, Pieper, U, Herzberg, O. | | Deposit date: | 1998-02-09 | | Release date: | 1998-05-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of the omega-loop in the activity, substrate specificity, and structure of class A beta-lactamase.
Biochemistry, 37, 1998
|
|
1PQ4
 
 | | Crystal structure of ZnuA | | Descriptor: | ZINC ION, periplasmic binding protein component of an ABC type zinc uptake transporter | | Authors: | Banerjee, S, Wei, B, Bhattacharyya-Pakrasi, M, Pakrasi, H.B, Smith, T.J. | | Deposit date: | 2003-06-17 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Determinants of Metal Specificity in the Zinc Transport Protein ZnuA from Synechocystis 6803.
J.Mol.Biol., 333, 2003
|
|
