3ZOA
 
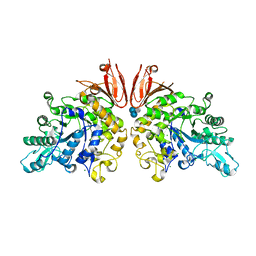 | | The structure of Trehalose Synthase (TreS) of Mycobacterium smegmatis in complex with acarbose | | 分子名称: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Caner, S, Nguyen, N, Aguda, A, Zhang, R, Pan, Y.T, Withers, S.G, Brayer, G.D. | | 登録日 | 2013-02-21 | | 公開日 | 2013-07-17 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | The Structure of the Mycobacterium Smegmatis Trehalose Synthase Reveals an Unusual Active Site Configuration and Acarbose-Binding Mode.
Glycobiology, 23, 2013
|
|
4NLE
 
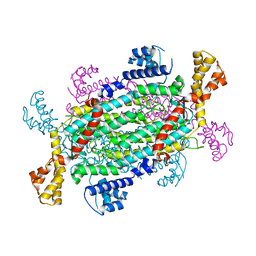 | |
4AJM
 
 | | Development of a plate-based optical biosensor methodology to identify PDE10 fragment inhibitors | | 分子名称: | 3-AMINO-6-FLUORO-2-[4-(2-METHYLPYRIDIN-4-YL)PHENYL]-N-(METHYLSULFONYL)QUINOLINE-4-CARBOXAMIDE, CAMP AND CAMP-INHIBITED CGMP 3', 5'-CYCLIC PHOSPHODIESTERASE 10A, ... | | 著者 | Geschwindner, S, Johansson, P, Spadola, L, Akerud, T, Back, E, Hillertz, P, Horsefeld, R, Scott, C, Spear, N, Tian, G, Tigerstrom, A, Aharony, D, Albert, J.S. | | 登録日 | 2012-02-16 | | 公開日 | 2013-03-06 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Development of a Plate-Based Optical Biosensor Methodology to Identify Pde10 Fragment Inhibitors
To be Published
|
|
3ZO9
 
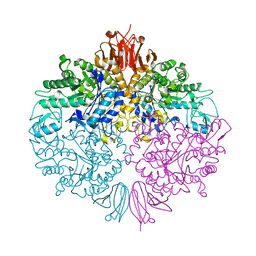 | | The structure of Trehalose Synthase (TreS) of Mycobacterium smegmatis | | 分子名称: | CALCIUM ION, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Caner, S, Nguyen, N, Aguda, A, Zhang, R, Pan, Y.T, Withers, S.G, Brayer, G.D. | | 登録日 | 2013-02-21 | | 公開日 | 2013-07-17 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | The Structure of the Mycobacterium Smegmatis Trehalose Synthase Reveals an Unusual Active Site Configuration and Acarbose-Binding Mode.
Glycobiology, 23, 2013
|
|
4B1R
 
 | |
4MTB
 
 | |
4NSL
 
 | |
2JQZ
 
 | | Solution Structure of the C2 domain of human Smurf2 | | 分子名称: | E3 ubiquitin-protein ligase SMURF2 | | 著者 | Wiesner, S, Ogunjimi, A.A, Wang, H, Rotin, D, Sicheri, F, Wrana, J.L, Forman-Kay, J.D. | | 登録日 | 2007-06-15 | | 公開日 | 2007-09-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Autoinhibition of the HECT-Type Ubiquitin Ligase Smurf2 through Its C2 Domain
Cell(Cambridge,Mass.), 130, 2007
|
|
2JCQ
 
 | | The hyaluronan binding domain of murine CD44 in a Type A complex with an HA 8-mer | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD44 ANTIGEN, GLYCEROL | | 著者 | Banerji, S, Wright, A.J, Noble, M.E.M, Mahoney, D.J, Campbell, I.D, Day, A.J, Jackson, D.G. | | 登録日 | 2007-01-03 | | 公開日 | 2007-01-30 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Structures of the Cd44-Hyaluronan Complex Provide Insight Into a Fundamental Carbohydrate-Protein Interaction.
Nat.Struct.Mol.Biol., 14, 2008
|
|
2JCP
 
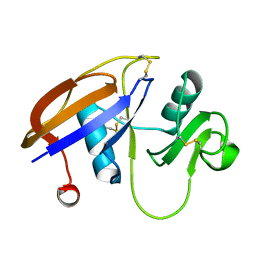 | | The hyaluronan binding domain of murine CD44 | | 分子名称: | CD44 ANTIGEN | | 著者 | Banerji, S, Wright, A.J, Noble, M.E.M, Mahoney, D.J, Campbell, I.D, Day, A.J, Jackson, D.G. | | 登録日 | 2007-01-03 | | 公開日 | 2007-01-30 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Structures of the Cd44-Hyaluronan Complex Provide Insight Into a Fundamental Carbohydrate-Protein Interaction.
Nat.Struct.Mol.Biol., 14, 2008
|
|
2K44
 
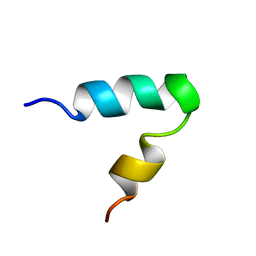 | |
2JCR
 
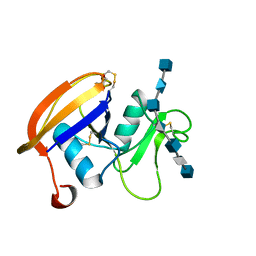 | | The hyaluronan binding domain of murine CD44 in a Type B complex with an HA 8-mer | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD44 ANTIGEN, GLYCEROL | | 著者 | Banerji, S, Wright, A.J, Noble, M.E.M, Mahoney, D.J, Campbell, I.D, Day, A.J, Jackson, D.G. | | 登録日 | 2007-01-03 | | 公開日 | 2007-01-30 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structures of the Cd44-Hyaluronan Complex Provide Insight Into a Fundamental Carbohydrate-Protein Interaction.
Nat.Struct.Mol.Biol., 14, 2008
|
|
2L9G
 
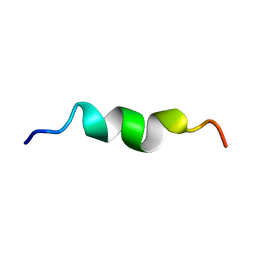 | |
6E5C
 
 | | Solution NMR structure of a de novo designed double-stranded beta-helix | | 分子名称: | De novo beta protein | | 著者 | Marcos, E, Chidyausiku, T.M, McShan, A, Evangelidis, T, Nerli, S, Sgourakis, N, Tripsianes, K, Baker, D. | | 登録日 | 2018-07-19 | | 公開日 | 2018-11-07 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | De novo design of a non-local beta-sheet protein with high stability and accuracy.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5WOX
 
 | |
5WOZ
 
 | |
1HMA
 
 | | THE SOLUTION STRUCTURE AND DYNAMICS OF THE DNA BINDING DOMAIN OF HMG-D FROM DROSOPHILA MELANOGASTER | | 分子名称: | HMG-D | | 著者 | Jones, D.N.M, Searles, M.A, Shaw, G.L, Churchill, M.E.A, Ner, S.S, Keeler, J, Travers, A.A, Neuhaus, D. | | 登録日 | 1994-05-12 | | 公開日 | 1994-07-31 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure and dynamics of the DNA-binding domain of HMG-D from Drosophila melanogaster.
Structure, 2, 1994
|
|
7RY6
 
 | | Solution NMR structural bundle of the first cyclization domain from yersiniabactin synthetase (Cy1) impacted by dynamics | | 分子名称: | HMWP2 nonribosomal peptide synthetase | | 著者 | Kancherla, A.K, Mishra, S.H, Marincin, K.A, Nerli, S, Sgourakis, N.G, Dowling, D.P, Bouvignies, G, Frueh, D.P. | | 登録日 | 2021-08-24 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Global protein dynamics as communication sensors in peptide synthetase domains.
Sci Adv, 8, 2022
|
|
4UIR
 
 | | Structure of oleate hydratase from Elizabethkingia meningoseptica | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, HEXAETHYLENE GLYCOL, OLEATE HYDRATASE, ... | | 著者 | Pavkov-Keller, T, Hromic, A, Engleder, M, Emmerstorfer, A, Steinkellner, G, Schrempf, S, Wriessnegger, T, Leitner, E, Strohmeier, G.A, Kaluzna, I, Mink, D, Schuermann, M, Wallner, S, Macheroux, P, Pichler, H, Gruber, K. | | 登録日 | 2015-04-02 | | 公開日 | 2015-07-01 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structure-Based Mechanism of Oleate Hydratase from Elizabethkingia Meningoseptica.
Chembiochem, 16, 2015
|
|
6O0I
 
 | | NMR ensemble of computationally designed protein XAA | | 分子名称: | Design construct XAA | | 著者 | Wei, K.Y, Moschidi, D, Nerli, S, Sgourakis, N, Baker, D. | | 登録日 | 2019-02-16 | | 公開日 | 2020-04-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Computational design of closely related proteins that adopt two well-defined but structurally divergent folds.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6O0C
 
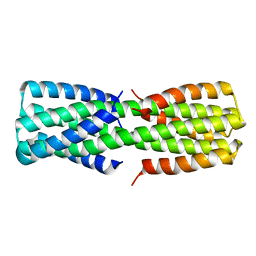 | | NMR ensemble of computationally designed protein XAA_GVDQ mutant M4L | | 分子名称: | Design construct XAA_GVDQ mutant M4L | | 著者 | Wei, K.Y, Moschidi, D, Nerli, S, Sgourakis, N, Baker, D. | | 登録日 | 2019-02-15 | | 公開日 | 2020-04-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Computational design of closely related proteins that adopt two well-defined but structurally divergent folds.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5WOT
 
 | |
5WOY
 
 | |
6E6M
 
 | | Crystal structure of human cellular retinol-binding protein 1 in complex with cannabidiorcin (CBDO) | | 分子名称: | (1'R,2'R)-4,5'-dimethyl-2'-(prop-1-en-2-yl)-1',2',3',4'-tetrahydro[1,1'-biphenyl]-2,6-diol, Retinol-binding protein 1 | | 著者 | Silvaroli, J.A, Horwitz, S, Banerjee, S, Kiser, P.D, Golczak, M. | | 登録日 | 2018-07-25 | | 公開日 | 2019-02-13 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Abnormal Cannabidiol Modulates Vitamin A Metabolism by Acting as a Competitive Inhibitor of CRBP1.
Acs Chem.Biol., 14, 2019
|
|
6OSW
 
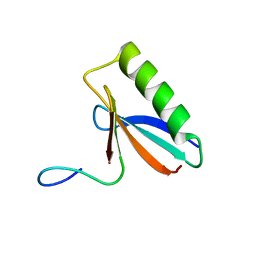 | | An order-to-disorder structural switch activates the FoxM1 transcription factor | | 分子名称: | Forkhead box M1 | | 著者 | Marceau, A.H, Rubin, S.M, Nerli, S, McShane, A.C, Sgourakis, N.G. | | 登録日 | 2019-05-02 | | 公開日 | 2019-05-29 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | An order-to-disorder structural switch activates the FoxM1 transcription factor.
Elife, 8, 2019
|
|
