5A6Z
 
 | | Structure of the LecB lectin from Pseudomonas aeruginosa strain PA14 in complex with lewis a | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, LECB, ... | | Authors: | Sommer, R, Wagner, S, Varrot, A, Khaledi, A, Haussler, S, Imberty, A, Titz, A. | | Deposit date: | 2015-07-02 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The virulence factor LecB varies in clinical isolates: consequences for ligand binding and drug discovery.
Chem Sci, 7, 2016
|
|
5I7E
 
 | |
4O6N
 
 | | Structure of AF2299, a CDP-alcohol phosphotransferase (CDP-bound) | | Descriptor: | AF2299, a CDP-alcohol phosphotransferase, CALCIUM ION, ... | | Authors: | Clarke, O.B, Sciara, G, Tomasek, D, Banerjee, S, Rajashankar, K.R, Shapiro, L, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2013-12-22 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for catalysis in a CDP-alcohol phosphotransferase.
Nat Commun, 5, 2014
|
|
4RRW
 
 | | Crystal Structure of Apo Murine Cyclooxygenase-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Prostaglandin G/H synthase 2, ... | | Authors: | Xu, S, Blobaum, A.L, Banerjee, S, Marnett, L.J. | | Deposit date: | 2014-11-06 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.569 Å) | | Cite: | Action at a Distance: MUTATIONS OF PERIPHERAL RESIDUES TRANSFORM RAPID REVERSIBLE INHIBITORS TO SLOW, TIGHT BINDERS OF CYCLOOXYGENASE-2.
J.Biol.Chem., 290, 2015
|
|
4M10
 
 | | Crystal Structure of Murine Cyclooxygenase-2 Complex with Isoxicam | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-hydroxy-2-methyl-N-(5-methyl-1,2-oxazol-3-yl)-2H-1,2-benzothiazine-3-carboxamide 1,1-dioxide, ... | | Authors: | Xu, S, Hermanson, D.J, Banerjee, S, Ghebreelasie, K, Marnett, L.J. | | Deposit date: | 2013-08-02 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Oxicams Bind in a Novel Mode to the Cyclooxygenase Active Site via a Two-water-mediated H-bonding Network.
J.Biol.Chem., 289, 2014
|
|
4RS0
 
 | | Crystal Structure of Murine H90W Cyclooxygenase-2 Complexed with S-ibuprofen | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IBUPROFEN, ... | | Authors: | Xu, S, Blobaum, A.L, Banerjee, S, Marnett, L.J. | | Deposit date: | 2014-11-06 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.807 Å) | | Cite: | Action at a Distance: MUTATIONS OF PERIPHERAL RESIDUES TRANSFORM RAPID REVERSIBLE INHIBITORS TO SLOW, TIGHT BINDERS OF CYCLOOXYGENASE-2.
J.Biol.Chem., 290, 2015
|
|
4O1Z
 
 | | Crystal Structure of Ovine Cyclooxygenase-1 Complex with Meloxicam | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-hydroxy-2-methyl-N-(5-methyl-1,3-thiazol-2-yl)-2H-1,2-benzothiazine-3-carboxamide 1,1-dioxide, ... | | Authors: | Xu, S, Hermanson, D.J, Banerjee, S, Ghebreselasie, K, Clayton, G.M, Garavito, R.M, Marnett, L.J. | | Deposit date: | 2013-12-16 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Oxicams Bind in a Novel Mode to the Cyclooxygenase Active Site via a Two-water-mediated H-bonding Network.
J.Biol.Chem., 289, 2014
|
|
4RRZ
 
 | | Crystal Structure of Apo Murine H90W Cyclooxygenase-2 Complexed with Lumiracoxib | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Prostaglandin G/H synthase 2, ... | | Authors: | Xu, S, Blobaum, A.L, Banerjee, S, Marnett, L.J. | | Deposit date: | 2014-11-06 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.569 Å) | | Cite: | Action at a Distance: MUTATIONS OF PERIPHERAL RESIDUES TRANSFORM RAPID REVERSIBLE INHIBITORS TO SLOW, TIGHT BINDERS OF CYCLOOXYGENASE-2.
J.Biol.Chem., 290, 2015
|
|
4O6M
 
 | | Structure of AF2299, a CDP-alcohol phosphotransferase (CMP-bound) | | Descriptor: | AF2299, a CDP-alcohol phosphotransferase, CALCIUM ION, ... | | Authors: | Clarke, O.B, Sciara, G, Tomasek, D, Banerjee, S, Rajashankar, K.R, Shapiro, L, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2013-12-22 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structural basis for catalysis in a CDP-alcohol phosphotransferase.
Nat Commun, 5, 2014
|
|
4RRY
 
 | | Crystal Structure of Apo Murine H90W Cyclooxygenase-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Prostaglandin G/H synthase 2, ... | | Authors: | Xu, S, Blobaum, A.L, Banerjee, S, Marnett, L.J. | | Deposit date: | 2014-11-06 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.429 Å) | | Cite: | Action at a Distance: MUTATIONS OF PERIPHERAL RESIDUES TRANSFORM RAPID REVERSIBLE INHIBITORS TO SLOW, TIGHT BINDERS OF CYCLOOXYGENASE-2.
J.Biol.Chem., 290, 2015
|
|
4RUT
 
 | | crystal structure of murine cyclooxygenase-2 with 13-methyl-arachidonic Acid | | Descriptor: | (5Z,8Z,11Z,13S,14Z)-13-methylicosa-5,8,11,14-tetraenoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xu, S, Kudalkar, S.N, Banerjee, S, Makriyannis, A, Nikas, S.P, Marnett, L.J. | | Deposit date: | 2014-11-21 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | 13-methylarachidonic Acid is a positive allosteric modulator of endocannabinoid oxygenation by cyclooxygenase.
J.Biol.Chem., 290, 2015
|
|
4OTJ
 
 | | The complex of murine cyclooxygenase-2 with a conjugate of indomefathin and podophyllotoxin, N-{(succinylpodophyllotoxinyl)but-4-yl}-2-{1-(4-chlorobenzoyl)-5-methoxy-2-methyl-1H-indol-3-yl}acetamide | | Descriptor: | (5S,5aS,8aS,9S)-8-oxo-9-(3,4,5-trimethoxyphenyl)-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol-5-yl 4-{[4-({[1-(4-chlorobenzoyl)-5-methoxy-2-methyl-1H-indol-3-yl]acetyl}amino)butyl]amino}-4-oxobutanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xu, S, Uddin, M.J, Banerjee, S, Marnett, L.J. | | Deposit date: | 2014-02-13 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Antitumor Activity of Cytotoxic Cyclooxygenase-2 Inhibitors.
Acs Chem.Biol., 11, 2016
|
|
5A1A
 
 | | 2.2 A resolution cryo-EM structure of beta-galactosidase in complex with a cell-permeant inhibitor | | Descriptor: | 2-phenylethyl 1-thio-beta-D-galactopyranoside, BETA-GALACTOSIDASE, MAGNESIUM ION, ... | | Authors: | Bartesaghi, A, Merk, A, Banerjee, S, Matthies, D, Wu, X, Milne, J, Subramaniam, S. | | Deposit date: | 2015-04-29 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | 2.2 A Resolution Cryo-Em Structure of Beta-Galactosidase in Complex with a Cell-Permeant Inhibitor
Science, 348, 2015
|
|
6WMV
 
 | | Structure of a phosphatidylinositol-phosphate synthase (PIPS) from Mycobacterium kansasii with evidence of substrate binding | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3,3',3''-phosphanetriyltripropanoic acid, AfCTD-Phosphatidylinositol-phosphate synthase (PIPS) fusion, ... | | Authors: | Belcher Dufrisne, M, Jorge, C.D, Timoteo, C.G, Petrou, V.I, Ashraf, K.U, Banerjee, S, Clarke, O.B, Santos, H, Mancia, F. | | Deposit date: | 2020-04-21 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.142 Å) | | Cite: | Structural and Functional Characterization of Phosphatidylinositol-Phosphate Biosynthesis in Mycobacteria.
J.Mol.Biol., 432, 2020
|
|
8Q6S
 
 | | A carbohydrate esterase family 15 (CE15) glucuronoyl esterase from Phocaeicola vulgatus ATCC 8482 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, Putative acetyl xylan esterase, ... | | Authors: | Mazurkewich, S, Seveso, A, Banerjee, S, Lo Leggio, L, Larsbrink, J. | | Deposit date: | 2023-08-14 | | Release date: | 2023-12-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Polysaccharide utilization loci from Bacteroidota encode CE15 enzymes with possible roles in cleaving pectin-lignin bonds.
Appl.Environ.Microbiol., 90, 2024
|
|
6N47
 
 | | The structure of SB-2-204-tubulin complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-chloropyrido[3,2-d]pyrimidin-4-yl)-7-methoxy-3,4-dihydroquinoxalin-2(1H)-one, CALCIUM ION, ... | | Authors: | Arnst, K, Banerjee, S, Wang, Y, Li, W, Miller, D, Li, W. | | Deposit date: | 2018-11-17 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray Crystal Structure Guided Discovery and Antitumor Efficacy of Dihydroquinoxalinone as Potent Tubulin Polymerization Inhibitors.
Acs Chem.Biol., 14, 2019
|
|
4OTY
 
 | | Crystal structure of lumiracoxib bound to the apo-mouse-cyclooxygenase-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Prostaglandin G/H synthase 2, ... | | Authors: | Xu, S, Windsor, M.A, Banerjee, S, Marnett, L.J. | | Deposit date: | 2014-02-14 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.354 Å) | | Cite: | Exploring the molecular determinants of substrate-selective inhibition of cyclooxygenase-2 by lumiracoxib.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
1RFR
 
 | | NMR structure of the 30mer stemloop-D of coxsackieviral RNA | | Descriptor: | stemloop-D RNA of the 5'-cloverleaf of coxsackievirus B3 | | Authors: | Ohlenschlager, O, Wohnert, J, Bucci, E, Seitz, S, Hafner, S, Ramachandran, R, Zell, R, Gorlach, M. | | Deposit date: | 2003-11-10 | | Release date: | 2004-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of the stemloop D subdomain of coxsackievirus B3 cloverleaf
RNA and its interaction with the proteinase 3C.
STRUCTURE, 12, 2004
|
|
6WM5
 
 | | Structure of a phosphatidylinositol-phosphate synthase (PIPS) from Mycobacterium kansasii | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, 3,3',3''-phosphanetriyltripropanoic acid, ... | | Authors: | Belcher Dufrisne, M, Jorge, C.D, Timoteo, C.G, Petrou, V.I, Ashraf, K.U, Banerjee, S, Clarke, O.B, Santos, H, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2020-04-20 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.961 Å) | | Cite: | Structural and Functional Characterization of Phosphatidylinositol-Phosphate Biosynthesis in Mycobacteria.
J.Mol.Biol., 432, 2020
|
|
6WQX
 
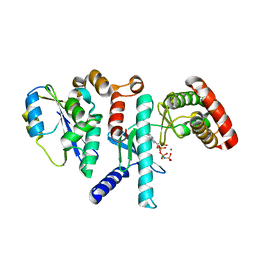 | | Human PRPK-TPRKB complex | | Descriptor: | EKC/KEOPS complex subunit TP53RK, EKC/KEOPS complex subunit TPRKB, MAGNESIUM ION, ... | | Authors: | Li, J, Ma, X.L, Banerjee, S, Dong, Z.G. | | Deposit date: | 2020-04-29 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Crystal structure of the human PRPK-TPRKB complex.
Commun Biol, 4, 2021
|
|
6X6O
 
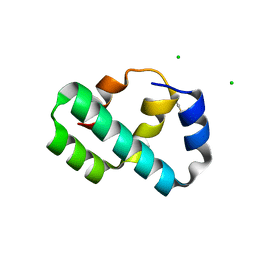 | | Crystal structure of T4 protein Spackle as determined by native SAD phasing | | Descriptor: | CHLORIDE ION, Protein spackle | | Authors: | Shi, K, Kurniawan, F, Banerjee, S, Moeller, N.H, Aihara, H. | | Deposit date: | 2020-05-28 | | Release date: | 2020-09-16 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal structure of bacteriophage T4 Spackle as determined by native SAD phasing.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
4QC9
 
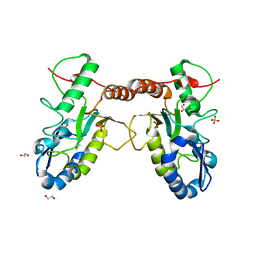 | | Crystal structure of Vaccinia virus uracil-DNA glycosylase mutant 3GD4 | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Uracil-DNA glycosylase | | Authors: | Sartmatova, D, Nash, T, Schormann, N, Nuth, M, Ricciardi, R, Banerjee, S, Chattopadhyay, D. | | Deposit date: | 2014-05-09 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.259 Å) | | Cite: | Crystal structure of Vaccinia virus uracil-DNA glycosylase mutant 3GD4
To be Published
|
|
3S96
 
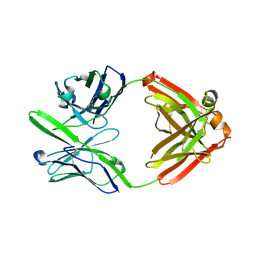 | | Crystal structure of 3B5H10 | | Descriptor: | 3B5H10 FAB heavy chain, 3B5H10 FAB light chain | | Authors: | Weisgraber, K, Peters-Libeu, C, Rutenber, E, Newhouse, Y, Finkbeiner, S. | | Deposit date: | 2011-05-31 | | Release date: | 2012-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Disease-associated polyglutamine stretches in monomeric huntingtin adopt a compact structure.
J.Mol.Biol., 421, 2012
|
|
5ILO
 
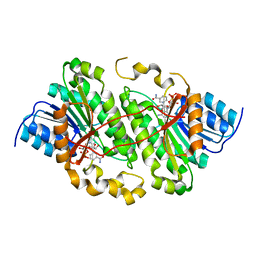 | | Crystal structure of photoreceptor dehydrogenase from Drosophila melanogaster | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Photoreceptor dehydrogenase, isoform C | | Authors: | Hofmann, L, Tsybovsky, Y, Banerjee, S. | | Deposit date: | 2016-03-04 | | Release date: | 2016-11-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural Insights into the Drosophila melanogaster Retinol Dehydrogenase, a Member of the Short-Chain Dehydrogenase/Reductase Family.
Biochemistry, 55, 2016
|
|
5TD5
 
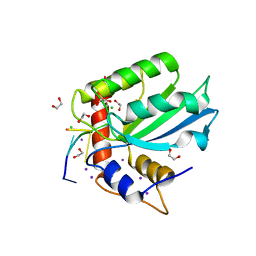 | | Crystal Structure of Human APOBEC3B variant complexed with ssDNA | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(P*TP*TP*CP*AP*T)-3'), ... | | Authors: | Shi, K, Banerjee, S, Kurahashi, K, Aihara, H. | | Deposit date: | 2016-09-16 | | Release date: | 2016-12-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.718 Å) | | Cite: | Structural basis for targeted DNA cytosine deamination and mutagenesis by APOBEC3A and APOBEC3B.
Nat. Struct. Mol. Biol., 24, 2017
|
|
