6W37
 
 | |
4GIQ
 
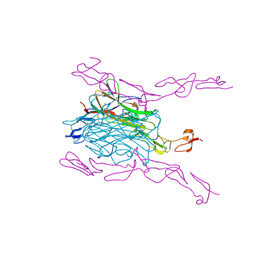 | | Crystal Structure of mouse RANK bound to RANKL | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, SODIUM ION, ... | | Authors: | Nelson, C.A, Wang, M.W.-H, Fremont, D.H. | | Deposit date: | 2012-08-08 | | Release date: | 2012-10-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | RANKL Employs Distinct Binding Modes to Engage RANK and the Osteoprotegerin Decoy Receptor.
Structure, 20, 2012
|
|
1XAK
 
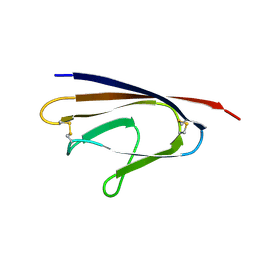 | |
1ZOX
 
 | |
6E47
 
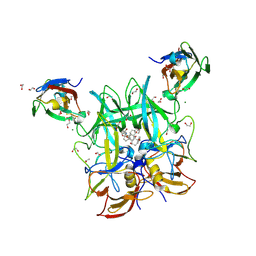 | |
6E48
 
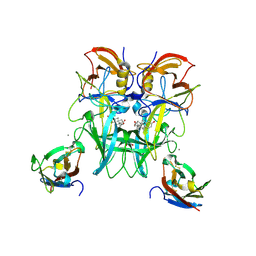 | | Crystal Structure of the Murine Norovirus VP1 P domain in complex with the CD300lf Receptor and Lithocholic Acid | | Descriptor: | (3beta,5beta,14beta,17alpha)-3-hydroxycholan-24-oic acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Nelson, C.A, Fremont, D.H. | | Deposit date: | 2018-07-16 | | Release date: | 2018-09-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural basis for murine norovirus engagement of bile acids and the CD300lf receptor.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6C6Q
 
 | |
6C74
 
 | |
5FFL
 
 | |
4E4D
 
 | | Crystal structure of mouse RANKL-OPG complex | | Descriptor: | CHLORIDE ION, Tumor necrosis factor ligand superfamily member 11, soluble form, ... | | Authors: | Nelson, C.A, Fremont, D.H. | | Deposit date: | 2012-03-12 | | Release date: | 2012-10-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | RANKL Employs Distinct Binding Modes to Engage RANK and the Osteoprotegerin Decoy Receptor.
Structure, 20, 2012
|
|
1XAU
 
 | |
3IRC
 
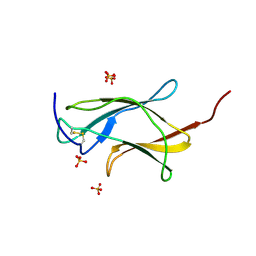 | | Crystal structure analysis of dengue-1 envelope protein domain III | | Descriptor: | ENVELOPE PROTEIN, SULFATE ION | | Authors: | Nelson, C.A, Kim, T, Warren, J.T, Chruszcz, M, Minor, W, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-21 | | Release date: | 2009-09-29 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure Analysis of the Dengue-1 Envelope Protein Domain III
To be Published
|
|
1JTZ
 
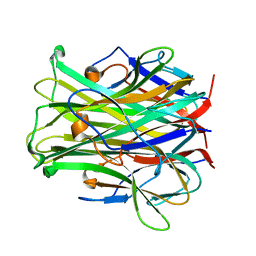 | | CRYSTAL STRUCTURE OF TRANCE/RANKL CYTOKINE. | | Descriptor: | TUMOR NECROSIS FACTOR LIGAND SUPERFAMILY MEMBER 11 | | Authors: | Nelson, C.A, Fremont, D.H. | | Deposit date: | 2001-08-23 | | Release date: | 2001-09-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the TRANCE/RANKL cytokine reveals determinants of receptor-ligand specificity
J.Clin.Invest., 108, 2001
|
|
3GK8
 
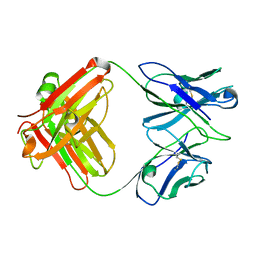 | | X-ray crystal structure of the Fab from MAb 14, mouse antibody against Canine Parvovirus | | Descriptor: | Fab 14 Heavy Chain, Fab 14 Light Chain | | Authors: | Hafenstein, S, Bowman, V, Sun, T, Nelson, C, Palermo, L, Chipman, P, Battisti, A, Parrish, C. | | Deposit date: | 2009-03-10 | | Release date: | 2009-06-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural comparison of different antibodies interacting with parvovirus capsids.
J.Virol., 83, 2009
|
|
1MQB
 
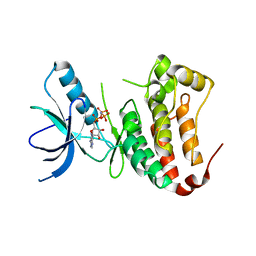 | | Crystal Structure of Ephrin A2 (ephA2) Receptor Protein Kinase | | Descriptor: | Ephrin type-A receptor 2, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Nowakowski, J, Cronin, C.N, McRee, D.E, Knuth, M.W, Nelson, C, Pavletich, N, Rogers, J, Sang, B.C, Scheibe, D.N, Swanson, R.V, Thompson, D.A. | | Deposit date: | 2002-09-16 | | Release date: | 2003-09-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of the Cancer Related Aurora-A, FAK and EphA2 Protein Kinases from Nanovolume Crystallography
Structure, 10, 2003
|
|
1MQ4
 
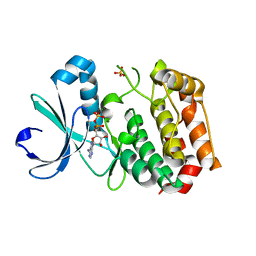 | | Crystal Structure of Aurora-A Protein Kinase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AURORA-RELATED KINASE 1, MAGNESIUM ION, ... | | Authors: | Nowakowski, J, Cronin, C.N, McRee, D.E, Knuth, M.W, Nelson, C, Pavletich, N.P, Rodgers, J, Sang, B.-C, Scheibe, D.N, Swanson, R.V, Thompson, D.A. | | Deposit date: | 2002-09-13 | | Release date: | 2003-09-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of the Cancer-Related Aurora-A, FAK and EphA2 Protein Kinases from Nanovolume Crystallography
Structure, 10, 2002
|
|
4FFE
 
 | | The structure of cowpox virus CPXV018 (OMCP) | | Descriptor: | CPXV018 protein | | Authors: | Lazear, E, Peterson, L.W, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-06-01 | | Release date: | 2012-06-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of the Cowpox Virus-Encoded NKG2D Ligand OMCP.
J.Virol., 87, 2013
|
|
4FG0
 
 | |
4FFY
 
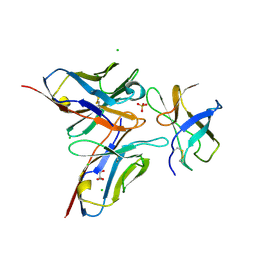 | | Crystal structure of DENV1-E111 single chain variable fragment bound to DENV-1 DIII, strain 16007. | | Descriptor: | CHLORIDE ION, DENV1-E111 single chain variable fragment (heavy chain), DENV1-E111 single chain variable fragment (light chain), ... | | Authors: | Austin, S.K, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-06-01 | | Release date: | 2012-06-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of Differential Neutralization of DENV-1 Genotypes by an Antibody that Recognizes a Cryptic Epitope.
Plos Pathog., 8, 2012
|
|
4FFZ
 
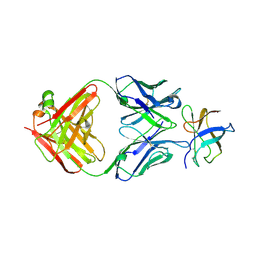 | | Crystal Structure of DENV1-E111 fab fragment bound to DENV-1 DIII (Western Pacific-74 strain). | | Descriptor: | DENV1-E111 fab fragment (heavy chain), DENV1-E111 fab fragment (light chain), Envelope protein E | | Authors: | Austin, S.K, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-06-01 | | Release date: | 2012-06-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural Basis of Differential Neutralization of DENV-1 Genotypes by an Antibody that Recognizes a Cryptic Epitope.
Plos Pathog., 8, 2012
|
|
8FXW
 
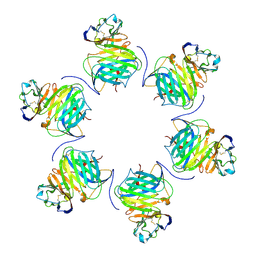 | | Cryo-EM structure of cowpox virus M2 in complex with human B7.1 (hexameric ring) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CPXV040 protein, T-lymphocyte activation antigen CD80 | | Authors: | Elliott, J.I, Adams, L.J, Nelson, C.A, Fremont, D.H, CSBID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2023-01-25 | | Release date: | 2025-02-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure basis of poxvirus mediated sabotage of T-cell costimulation
To Be Published
|
|
8FXZ
 
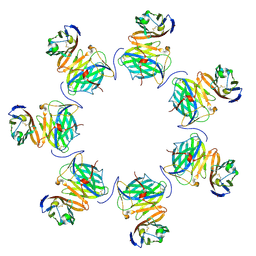 | | Cryo-EM structure of cowpox virus M2 in complex with human B7.1 (heptameric ring) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CPXV040 protein, T-lymphocyte activation antigen CD80 | | Authors: | Elliott, J.I, Adams, L.J, Nelson, C.A, Fremont, D.H, CSBID | | Deposit date: | 2023-01-25 | | Release date: | 2025-02-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structure basis of poxvirus mediated sabotage of T-cell costimulation
To Be Published
|
|
8FXY
 
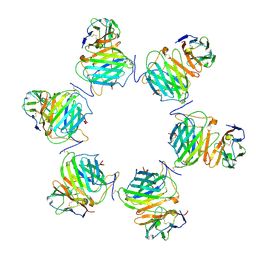 | | Cryo-EM structure of cowpox virus M2 in complex with human B7.2 (hexameric ring) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CPXV040 protein, T-lymphocyte activation antigen CD86 | | Authors: | Elliott, J.I, Adams, L.J, Nelson, C.A, Fremont, D.H, CSBID | | Deposit date: | 2023-01-25 | | Release date: | 2025-02-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structure basis of poxvirus mediated sabotage of T-cell costimulation
To Be Published
|
|
8FXX
 
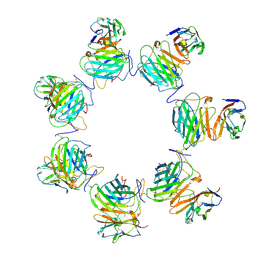 | | Cryo-EM structure of cowpox virus M2 in complex with human B7.2 (heptameric ring) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CPXV040 protein, T-lymphocyte activation antigen CD86 | | Authors: | Elliott, J.I, Adams, L.J, Nelson, C.A, Fremont, D.H, CSBID | | Deposit date: | 2023-01-25 | | Release date: | 2025-02-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structure basis of poxvirus mediated sabotage of T-cell costimulation
To Be Published
|
|
3M5R
 
 | |
