4FGS
 
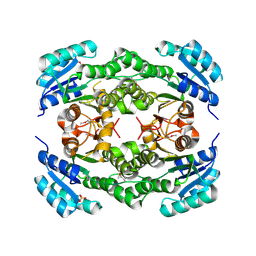 | | Crystal structure of a probable dehydrogenase protein | | Descriptor: | Probable dehydrogenase protein, SULFATE ION | | Authors: | Eswaramoorthy, S, Rice, S, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-06-04 | | Release date: | 2012-08-15 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of a probable dehydrogenase protein
To be Published
|
|
4FB5
 
 | |
6QMP
 
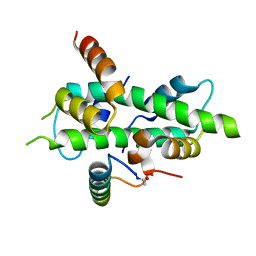 | | NF-YB/C Heterodimer in Complex with NF-YA Peptide | | Descriptor: | Nuclear transcription factor Y subunit alpha, Nuclear transcription factor Y subunit beta, Nuclear transcription factor Y subunit gamma | | Authors: | Kiehstaller, S, Jeganathan, S, Pearce, N.M, Wendt, M, Grossmann, T.N, Hennig, S. | | Deposit date: | 2019-02-08 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Constrained Peptides with Fine-Tuned Flexibility Inhibit NF-Y Transcription Factor Assembly.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6QMS
 
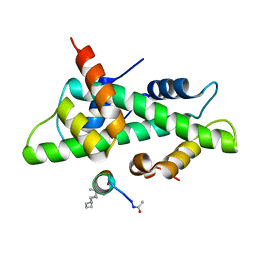 | | NF-YB/C Heterodimer in Complex with NF-YA-derived Peptide Stabilized with C11-Hydrocarbon Linker | | Descriptor: | Nuclear transcription factor Y subunit alpha, Nuclear transcription factor Y subunit beta, Nuclear transcription factor Y subunit gamma | | Authors: | Kiehstaller, S, Jeganathan, S, Pearce, N.M, Wendt, M, Grossmann, T.N, Hennig, S. | | Deposit date: | 2019-02-08 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Constrained Peptides with Fine-Tuned Flexibility Inhibit NF-Y Transcription Factor Assembly.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
3UMA
 
 | | Crystal structure of a hypothetical peroxiredoxin protein frm Sinorhizobium meliloti | | Descriptor: | Hypothetical peroxiredoxin protein, SULFATE ION | | Authors: | Eswaramoorthy, S, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-11-12 | | Release date: | 2011-11-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a hypothetical peroxiredoxin protein from Sinorhizobium meliloti
To be Published
|
|
3UP8
 
 | | Crystal structure of a putative 2,5-diketo-D-gluconic acid reductase B | | Descriptor: | ACETATE ION, Putative 2,5-diketo-D-gluconic acid reductase B | | Authors: | Eswaramoorthy, S, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-11-17 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of a putative 2,5-diketo-D-gluconic acid reductase B
To be Published
|
|
3C9G
 
 | |
3BPD
 
 | |
5H2U
 
 | | Crystal structure of PTK6 Kinase Domain complexed with Dasatinib | | Descriptor: | GLYCEROL, N-(2-CHLORO-6-METHYLPHENYL)-2-({6-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]-2-METHYLPYRIMIDIN-4-YL}AMINO)-1,3-THIAZOLE-5-CARBOXAMIDE, Protein-tyrosine kinase 6 | | Authors: | Thakur, M.K, Birudukota, S, Swaminathan, S, Tyagi, R, Gosu, R. | | Deposit date: | 2016-10-18 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Co-crystal structures of PTK6: With Dasatinib at 2.24 angstrom , with novel imidazo[1,2-a]pyrazin-8-amine derivative inhibitor at 1.70 angstrom resolution
Biochem. Biophys. Res. Commun., 482, 2017
|
|
3CBN
 
 | |
3US8
 
 | | Crystal Structure of an isocitrate dehydrogenase from Sinorhizobium meliloti 1021 | | Descriptor: | Isocitrate dehydrogenase [NADP], SULFATE ION | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-11-23 | | Release date: | 2011-12-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of an isocitrate dehydrogenase from Sinorhizobium meliloti 1021
To be Published
|
|
3UXY
 
 | | The crystal structure of short chain dehydrogenase from Rhodobacter sphaeroides | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Short-chain dehydrogenase/reductase SDR | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-05 | | Release date: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | The crystal structure of short chain dehydrogenase from Rhodobacter sphaeroides
To be Published
|
|
3V2G
 
 | | Crystal structure of a dehydrogenase/reductase from Sinorhizobium meliloti 1021 | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase, SULFATE ION | | Authors: | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-12 | | Release date: | 2012-01-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a dehydrogenase/reductase from Sinorhizobium meliloti 1021
To be Published
|
|
3V2H
 
 | | The crystal structure of D-beta-hydroxybutyrate dehydrogenase from Sinorhizobium meliloti | | Descriptor: | D-beta-hydroxybutyrate dehydrogenase | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-12 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of D-beta-hydroxybutyrate dehydrogenase from Sinorhizobium meliloti
To be Published
|
|
3BWI
 
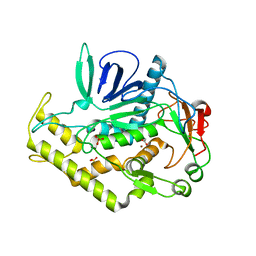 | | Crystal structure of the catalytic domain of botulinum neurotoxin serotype A with an acetate ion bound at the active site | | Descriptor: | ACETATE ION, Botulinum neurotoxin A light chain, SULFATE ION, ... | | Authors: | Kumaran, D, Rawat, R, Swaminathan, S. | | Deposit date: | 2008-01-09 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure- and Substrate-based Inhibitor Design for Clostridium botulinum Neurotoxin Serotype A
J.Biol.Chem., 283, 2008
|
|
3C88
 
 | |
3BT3
 
 | |
3UN1
 
 | | Crystal structure of an oxidoreductase from Sinorhizobium meliloti 1021 | | Descriptor: | PHOSPHATE ION, Probable oxidoreductase | | Authors: | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-11-15 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of an oxidoreductase from Sinorhizobium meliloti 1021
To be Published
|
|
3C8B
 
 | |
3C89
 
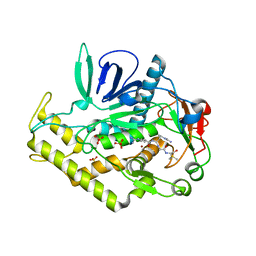 | |
3L3S
 
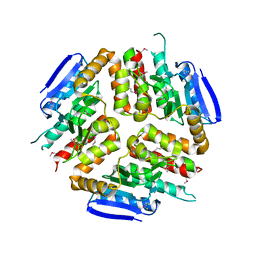 | |
3UOE
 
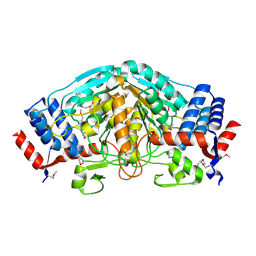 | | The crystal structure of dehydrogenase from Sinorhizobium meliloti | | Descriptor: | Dehydrogenase | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-11-16 | | Release date: | 2011-11-30 | | Method: | X-RAY DIFFRACTION (2.311 Å) | | Cite: | The crystal structure of dehydrogenase from Sinorhizobium meliloti
To be Published
|
|
3CIH
 
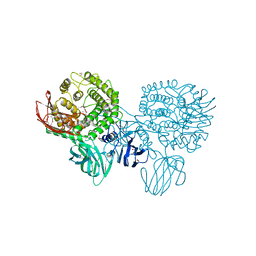 | | Crystal structure of a putative alpha-rhamnosidase from Bacteroides thetaiotaomicron | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Putative alpha-rhamnosidase | | Authors: | Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-11 | | Release date: | 2008-04-01 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of a putative alpha-rhamnosidase from Bacteroides thetaiotaomicron.
To be Published
|
|
3CLK
 
 | |
3CO4
 
 | |
