5JX7
 
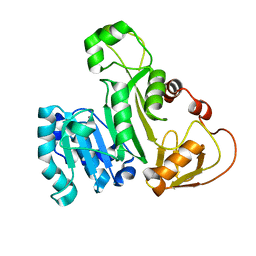 | | Cysteine mutant (C224A) structure of As (III) S-adenosyl methyltransferase | | Descriptor: | Arsenic methyltransferase, CALCIUM ION | | Authors: | Packianathan, C, Marapakala, K, Ajees, A.A, Kandavelu, P, Sankaran, B, Rosen, B.P. | | Deposit date: | 2016-05-12 | | Release date: | 2017-05-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cysteine mutant (C224A) structure of As (III) S-adenosyl methyltransferase
To be Published
|
|
7JWX
 
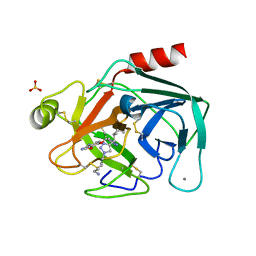 | | Crystal Structure of Trypsin Bound O-methyl Benzamidine | | Descriptor: | 4-[(1-{(1S,2S)-1-[1-(4-aminobutyl)-1H-1,2,3-triazol-4-yl]-2-methylbutyl}-1H-1,2,3-triazol-4-yl)methoxy]-3-methoxybenzene-1-carboximidamide, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Packianathan, C, Laganowsky, A. | | Deposit date: | 2020-08-26 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Small molecule peptidomimetic trypsin inhibitors: validation of an EKO binding mode, but with a twist.
Org.Biomol.Chem., 20, 2022
|
|
6CX6
 
 | |
4KW7
 
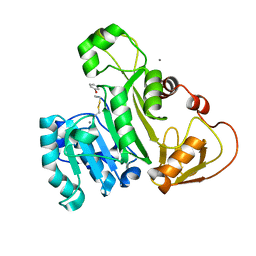 | | The structure of an As(III) S-adenosylmethionine methyltransferase with Phenylarsine oxide(PAO) | | Descriptor: | Arsenic methyltransferase, CALCIUM ION, Phenylarsine oxide | | Authors: | Packianathan, C, Marapakala, K, Ajees, A.A, Kandavelu, P, Rosen, B.P. | | Deposit date: | 2013-05-23 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A disulfide-bond cascade mechanism for arsenic(III) S-adenosylmethionine methyltransferase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5EG5
 
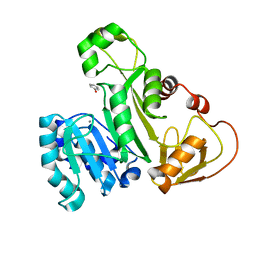 | |
4RSR
 
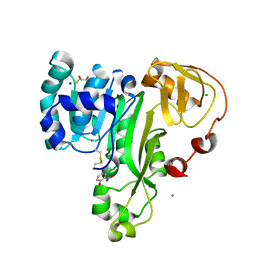 | | ArsM arsenic(III) S-adenosylmethionine methyltransferase with trivalent phenyl arsencial derivative-Roxarsone | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-arsanyl-2-nitrophenol, Arsenic methyltransferase, ... | | Authors: | Packianathan, C, Marapakala, K, Ajees, A.A, Kandavelu, P, Rosen, B.P. | | Deposit date: | 2014-11-10 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A disulfide-bond cascade mechanism for arsenic(III) S-adenosylmethionine methyltransferase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5EVJ
 
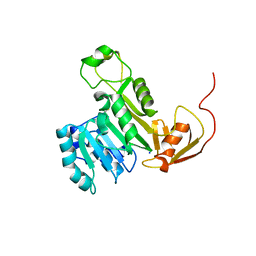 | | X-ray crystal structure of CrArsM, an arsenic (III) S-adenosylmethionine methyltransferase from Chlamydomonas reinhardtii | | Descriptor: | Arsenite methyltransferase, SODIUM ION | | Authors: | Packianathan, C, Kandavelu, P, Sankaran, B, Rosen, B.P. | | Deposit date: | 2015-11-19 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of CrArsM, an arsenic (III) S-adenosylmethionine methyltransferase from Chlamydomonas reinhardtii
To Be Published
|
|
3KXS
 
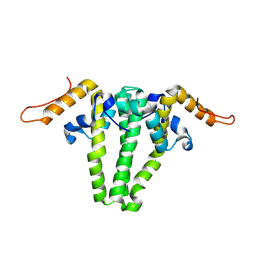 | |
2QLS
 
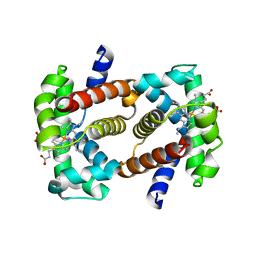 | | crystal structure of hemoglobin from dog (Canis familiaris) at 3.5 Angstrom resolution | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Packianathan, C, Sundaresan, S, Palani, K, Neeelagandan, K, Ponnuswamy, M.N. | | Deposit date: | 2007-07-13 | | Release date: | 2008-07-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Purification, Crystallization and Crystal structure analysis of hemoglobin from Dog (Canis familiaris)
To be Published
|
|
2QMB
 
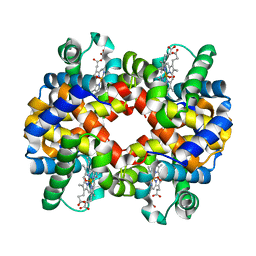 | |
2A8X
 
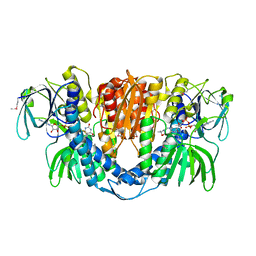 | | Crystal Structure of Lipoamide Dehydrogenase from Mycobacterium tuberculosis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Rajashankar, K.R, Bryk, R, Kniewel, R, Buglino, J.A, Nathan, C.F, Lima, C.D. | | Deposit date: | 2005-07-10 | | Release date: | 2005-08-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure and functional analysis of lipoamide dehydrogenase from Mycobacterium tuberculosis
J.Biol.Chem., 280, 2005
|
|
1KNC
 
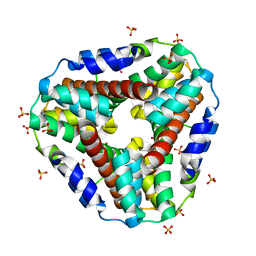 | | Structure of AhpD from Mycobacterium tuberculosis, a novel enzyme with thioredoxin-like activity. | | Descriptor: | AhpD protein, SULFATE ION | | Authors: | Bryk, R, Lima, C.D, Erdjument-Bromage, H, Tempst, P, Nathan, C. | | Deposit date: | 2001-12-18 | | Release date: | 2002-01-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metabolic enzymes of mycobacteria linked to antioxidant defense by a thioredoxin-like protein.
Science, 295, 2002
|
|
2FHG
 
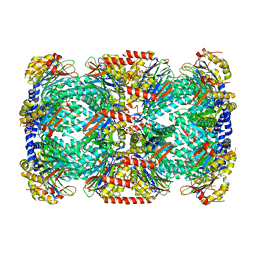 | | Crystal Structure of Mycobacterial Tuberculosis Proteasome | | Descriptor: | 20S proteasome, alpha and beta subunits, proteasome, ... | | Authors: | Hu, G, Lin, G, Wang, M, Dick, L, Xu, R.M, Nathan, C, Li, H. | | Deposit date: | 2005-12-23 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Structure of the Mycobacterium tuberculosis proteasome and mechanism of inhibition by a peptidyl boronate.
Mol.Microbiol., 59, 2006
|
|
5TS0
 
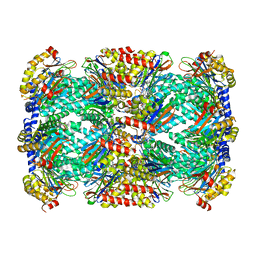 | | Structure of Mycobacterium tuberculosis proteasome in complex with N,C-capped dipeptide PKS2208 | | Descriptor: | (2S)-N-{(2S)-3-methoxy-1-[(naphthalen-1-ylmethyl)amino]-1-oxopropan-2-yl}-4-oxo-2-[(3-phenylpropanoyl)amino]-4-(1H-pyrrol-1-yl)butanamide (non-preferred name), Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Hsu, H.-C, Fan, H, Singh, P.K, Wang, R, Sukenick, G, Nathan, C, Lin, G, Li, H. | | Deposit date: | 2016-10-27 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.84679747 Å) | | Cite: | Structural Basis for the Species-Selective Binding of N,C-Capped Dipeptides to the Mycobacterium tuberculosis Proteasome.
Biochemistry, 56, 2017
|
|
5TRS
 
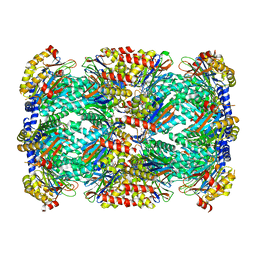 | | Structure of Mycobacterium tuberculosis proteasome in complex with N,C-capped dipeptide PKS2144 | | Descriptor: | N-tert-butoxy-N~2~-(5-methyl-1,2-oxazole-3-carbonyl)-L-asparaginyl-O-methyl-N-[(naphthalen-1-yl)methyl]-L-serinamide, Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Hsu, H.-C, Fan, H, Singh, P.K, Wang, R, Sukenick, G, Nathan, C, Lin, G, Li, H. | | Deposit date: | 2016-10-27 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.083567 Å) | | Cite: | Structural Basis for the Species-Selective Binding of N,C-Capped Dipeptides to the Mycobacterium tuberculosis Proteasome.
Biochemistry, 56, 2017
|
|
5TRY
 
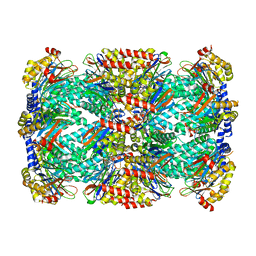 | | Structure of Mycobacterium tuberculosis proteasome in complex with N,C-capped dipeptide PKS2206 | | Descriptor: | (2~{S})-~{N}-[(2~{S})-3-methoxy-1-(naphthalen-1-ylmethylamino)-1-oxidanylidene-propan-2-yl]-4-oxidanylidene-2-(3-phenylpropanoylamino)-4-piperidin-1-yl-butanamide, Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Hsu, H.-C, Fan, H, Singh, P.K, Wang, R, Sukenick, G, Nathan, C, Lin, G, Li, H. | | Deposit date: | 2016-10-27 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.000008 Å) | | Cite: | Structural Basis for the Species-Selective Binding of N,C-Capped Dipeptides to the Mycobacterium tuberculosis Proteasome.
Biochemistry, 56, 2017
|
|
5TRR
 
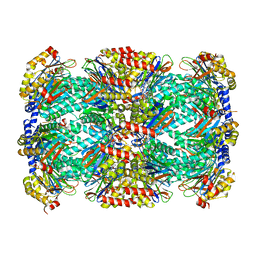 | | Structure of Mycobacterium tuberculosis proteasome in complex with N,C-capped dipeptide PKS2169 | | Descriptor: | N,N-diethyl-N~2~-(3-phenylpropanoyl)-L-asparaginyl-N-[(naphthalen-1-yl)methyl]-L-alaninamide, Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Hsu, H.-C, Fan, H, Singh, P.K, Wang, R, Sukenick, G, Nathan, C, Lin, G, Li, H. | | Deposit date: | 2016-10-27 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.103 Å) | | Cite: | Structural Basis for the Species-Selective Binding of N,C-Capped Dipeptides to the Mycobacterium tuberculosis Proteasome.
Biochemistry, 56, 2017
|
|
5TRG
 
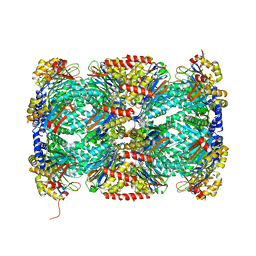 | | Structure of Mycobacterium tuberculosis proteasome in complex with N,C-capped dipeptide DPLG-2 | | Descriptor: | N,N-diethyl-N~2~-[(2E)-3-phenylprop-2-enoyl]-L-asparaginyl-4-fluoro-N-[(naphthalen-1-yl)methyl]-L-phenylalaninamide, Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Hsu, H.-C, Fan, H, Singh, R.K, Wang, R, Sukenick, G, Nathan, C, Lin, G, Li, H. | | Deposit date: | 2016-10-26 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Structural Basis for the Species-Selective Binding of N,C-Capped Dipeptides to the Mycobacterium tuberculosis Proteasome.
Biochemistry, 56, 2017
|
|
2AQ3
 
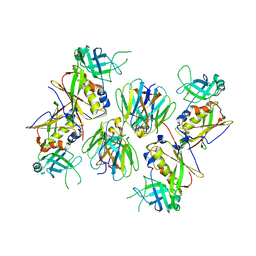 | | Crystal structure of T-cell receptor V beta domain variant complexed with superantigen SEC3 | | Descriptor: | Enterotoxin type C-3, T-cell receptor beta chain V | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-17 | | Release date: | 2006-03-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
2AQ2
 
 | | Crystal structure of T-cell receptor V beta domain variant complexed with superantigen SEC3 mutant | | Descriptor: | Enterotoxin type C-3, SODIUM ION, SULFATE ION, ... | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-17 | | Release date: | 2006-03-21 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
2AQ1
 
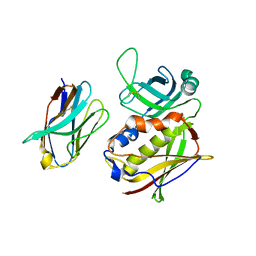 | | Crystal structure of T-cell receptor V beta domain variant complexed with superantigen SEC3 mutant | | Descriptor: | Enterotoxin type C-3, T-cell receptor beta chain V | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-17 | | Release date: | 2006-03-21 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
2APT
 
 | | Crystal Structure of the G17E/S54N/K66E/Q72H/E80V/L81S/T87S/G96V variant of the murine T cell receptor V beta 8.2 domain | | Descriptor: | MALONIC ACID, T-cell receptor beta chain V | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-16 | | Release date: | 2006-03-21 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
2APB
 
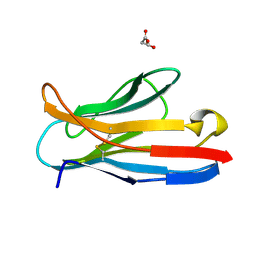 | | Crystal Structure of the S54N variant of murine T cell receptor Vbeta 8.2 domain | | Descriptor: | MALONIC ACID, T-cell receptor beta chain V | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-16 | | Release date: | 2006-03-21 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
2APX
 
 | | Crystal Structure of the G17E/A52V/S54N/K66E/Q72H/E80V/L81S/T87S/G96V variant of the murine T cell receptor V beta 8.2 domain | | Descriptor: | MALONIC ACID, T cell receptor beta chain V | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-16 | | Release date: | 2006-03-21 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
2APW
 
 | | Crystal Structure of the G17E/A52V/S54N/K66E/E80V/L81S/T87S/G96V variant of the murine T cell receptor V beta 8.2 domain | | Descriptor: | MALONIC ACID, T cell receptor beta chain V | | Authors: | Cho, S, Swaminathan, C.P, Yang, J, Kerzic, M.C, Guan, R, Kieke, M.C, Kranz, D.M, Mariuzza, R.A, Sundberg, E.J. | | Deposit date: | 2005-08-16 | | Release date: | 2006-03-21 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of affinity maturation and intramolecular cooperativity in a protein-protein interaction.
Structure, 13, 2005
|
|
