2PLG
 
 | |
2POZ
 
 | |
3M1Y
 
 | |
2Q09
 
 | | Crystal structure of Imidazolonepropionase from environmental sample with bound inhibitor 3-(2,5-Dioxo-imidazolidin-4-yl)-propionic acid | | Descriptor: | 3-[(4S)-2,5-DIOXOIMIDAZOLIDIN-4-YL]PROPANOIC ACID, FE (III) ION, Imidazolonepropionase | | Authors: | Tyagi, R, Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-05-21 | | Release date: | 2007-06-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A common catalytic mechanism for proteins of the HutI family.
Biochemistry, 47, 2008
|
|
3US8
 
 | | Crystal Structure of an isocitrate dehydrogenase from Sinorhizobium meliloti 1021 | | Descriptor: | Isocitrate dehydrogenase [NADP], SULFATE ION | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-11-23 | | Release date: | 2011-12-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of an isocitrate dehydrogenase from Sinorhizobium meliloti 1021
To be Published
|
|
2N3O
 
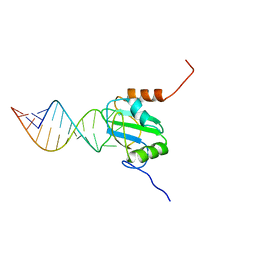 | | Structure of PTB RRM1(41-163) bound to an RNA stemloop containing a structured loop derived from viral internal ribosomal entry site RNA | | Descriptor: | Polypyrimidine tract-binding protein 1, RNA (5'-R(*GP*GP*GP*AP*CP*CP*UP*GP*GP*UP*CP*UP*UP*UP*CP*CP*AP*GP*GP*UP*CP*CP*C)-3') | | Authors: | Maris, C, Jayne, S.F, Damberger, F.F, Ravindranathan, S, Allain, F.H.-T. | | Deposit date: | 2015-06-08 | | Release date: | 2016-08-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | C-terminal helix folding upon pyrimidine-rich hairpin binding to PTB RRM1. Implications for PTB function in Encephalomyocarditis virus IRES activity.
To be Published
|
|
3DDA
 
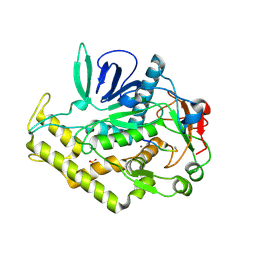 | |
3DDB
 
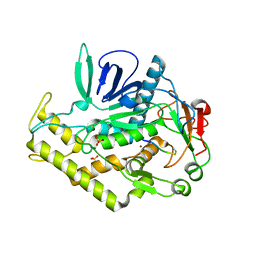 | |
3MHD
 
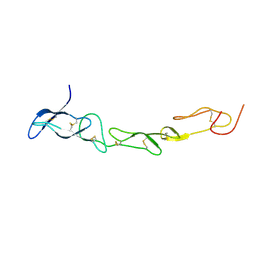 | | Crystal structure of DCR3 | | Descriptor: | Tumor necrosis factor receptor superfamily member 6B | | Authors: | Zhan, C, Patskovsky, Y, Yan, Q, Li, Z, Ramagopal, U.A, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2010-04-07 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Decoy Strategies: The Structure of TL1A:DcR3 Complex.
Structure, 19, 2011
|
|
3M2P
 
 | |
3N53
 
 | |
4EN0
 
 | | Crystal structure of light | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Zhan, C, Liu, W, Patskovsky, Y, Ramagopal, U.A, Bonanno, J.B, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2012-04-12 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Mechanistic basis for functional promiscuity in the TNF and TNF receptor superfamilies: structure of the LIGHT:DcR3 assembly.
Structure, 22, 2014
|
|
3U4J
 
 | | Crystal structure of NAD-dependent aldehyde dehydrogenase from Sinorhizobium meliloti | | Descriptor: | CALCIUM ION, NAD-dependent aldehyde dehydrogenase | | Authors: | Eswaramoorthy, S, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-10-08 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of NAD-dependent aldehyde dehydrogenase from Sinorhizobium meliloti
To be Published
|
|
3T9P
 
 | | Crystal structure of a putative mandelate racemase/muconate lactonizing enzyme family protein from Roseovarius | | Descriptor: | FORMIC ACID, GLYCEROL, Mandelate racemase/muconate lactonizing enzyme family protein, ... | | Authors: | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-08-03 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of a putative mandelate racemase/muconate lactonizing enzyme family protein from Roseovarius
To be Published
|
|
2QVG
 
 | |
3DLI
 
 | |
3DZB
 
 | |
3KZG
 
 | |
2GV8
 
 | | Crystal structure of flavin-containing monooxygenase (FMO) from S.pombe and NADPH cofactor complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Eswaramoorthy, S, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-05-02 | | Release date: | 2006-06-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of action of a flavin-containing monooxygenase.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3ROS
 
 | |
1XD7
 
 | |
3NHM
 
 | |
2PHP
 
 | |
1Z2L
 
 | | Crystal structure of Allantoate-amidohydrolase from E.coli K12 in complex with substrate Allantoate | | Descriptor: | ALLANTOATE ION, Allantoate amidohydrolase, SULFATE ION, ... | | Authors: | Agarwal, R, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-03-08 | | Release date: | 2005-03-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural analysis of a ternary complex of allantoate amidohydrolase from Escherichia coli reveals its mechanics.
J.Mol.Biol., 368, 2007
|
|
3EHE
 
 | |
