6TLU
 
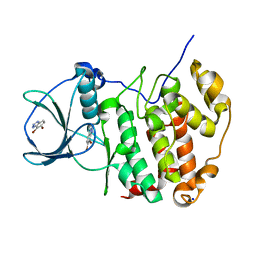 | | HUMAN CK2 KINASE ALPHA SUBUNIT IN COMPLEX WITH THE ATP-COMPETITIVE INHIBITOR 4,5-DIBROMOBENZOTRIAZOLE | | Descriptor: | 6,7-bis(bromanyl)-1~{H}-benzotriazole, Casein kinase II subunit alpha, SODIUM ION | | Authors: | Czapinska, H, Piasecka, A, Winiewska-Szajewska, M, Bochtler, M, Poznanski, J. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Halogen Atoms in the Protein-Ligand System. Structural and Thermodynamic Studies of the Binding of Bromobenzotriazoles by the Catalytic Subunit of Human Protein Kinase CK2.
J.Phys.Chem.B, 125, 2021
|
|
4O9K
 
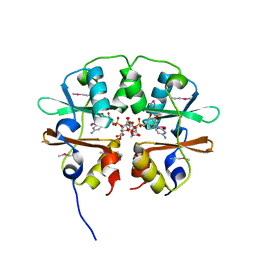 | | Crystal structure of the CBS pair of a putative D-arabinose 5-phosphate isomerase from Methylococcus capsulatus in complex with CMP-Kdo | | Descriptor: | Arabinose 5-phosphate isomerase, CYTIDINE 5'-MONOPHOSPHATE 3-DEOXY-BETA-D-GULO-OCT-2-ULO-PYRANOSONIC ACID, GLYCEROL | | Authors: | Shabalin, I.G, Cooper, D.R, Shumilin, I.A, Zimmerman, M.D, Majorek, K.A, Hammonds, J, Hillerich, B.S, Nawar, A, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-01-02 | | Release date: | 2014-01-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure and kinetic properties of D-arabinose 5-phosphate isomerase from Methylococcus capsulatus
To be Published
|
|
6YWN
 
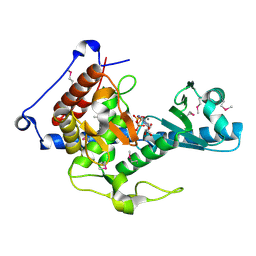 | | CutA in complex with CMPCPP | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, CALCIUM ION, CutA | | Authors: | Malik, D, Kobylecki, K, Krawczyk, P, Poznanski, J, Jakielaszek, A, Napiorkowska, A, Dziembowski, A, Tomecki, R, Nowotny, M. | | Deposit date: | 2020-04-29 | | Release date: | 2020-08-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and mechanism of CutA, RNA nucleotidyl transferase with an unusual preference for cytosine.
Nucleic Acids Res., 48, 2020
|
|
6YWP
 
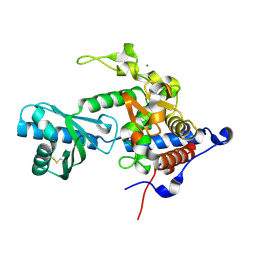 | | Structure of apo-CutA | | Descriptor: | CutA, MAGNESIUM ION | | Authors: | Malik, D, Kobylecki, K, Krawczyk, P, Poznanski, J, Jakielaszek, A, Napiorkowska, A, Dziembowski, A, Tomecki, R, Nowotny, M. | | Deposit date: | 2020-04-29 | | Release date: | 2020-08-05 | | Last modified: | 2020-09-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure and mechanism of CutA, RNA nucleotidyl transferase with an unusual preference for cytosine.
Nucleic Acids Res., 48, 2020
|
|
6YWO
 
 | | CutA in complex with A3 RNA | | Descriptor: | CHLORIDE ION, CutA, MAGNESIUM ION, ... | | Authors: | Malik, D, Kobylecki, K, Krawczyk, P, Poznanski, J, Jakielaszek, A, Napiorkowska, A, Dziembowski, A, Tomecki, R, Nowotny, M. | | Deposit date: | 2020-04-29 | | Release date: | 2020-08-05 | | Last modified: | 2020-09-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of CutA, RNA nucleotidyl transferase with an unusual preference for cytosine.
Nucleic Acids Res., 48, 2020
|
|
4N18
 
 | | Crystal structure of D-isomer specific 2-hydroxyacid dehydrogenase family protein from Klebsiella pneumoniae 342 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, CITRIC ACID, D-isomer specific 2-hydroxyacid dehydrogenase family protein | | Authors: | Bacal, P, Shabalin, I.G, Cooper, D.R, Majorek, K.A, Osinski, T, Hillerich, B.S, Hammonds, J, Nawar, A, Stead, M, Chowdhury, S, Gizzi, A, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-10-03 | | Release date: | 2013-10-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of D-isomer specific 2-hydroxyacid dehydrogenase family protein from Klebsiella pneumoniae 342
To be Published
|
|
3SNL
 
 | | Highly Potent, Selective, and Orally Active Phosphodiestarase 10A Inhibitors | | Descriptor: | 6-chloro-3,4-dimethyl-1-(3-methylpyridin-4-yl)-8-(trifluoromethyl)imidazo[1,5-a]quinoxaline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Malamas, M.S, Ni, Y, Erdei, J, Stange, H, Schindler, R, Lankau, H.-J, Grunwald, C, Fan, K.Y, Parris, K.D, Langen, B, Egerland, U, Hage, T, Marquis, K.L, Grauer, S, Brennan, J, Navarra, R, Graf, R, Harrison, B.L, Robichaud, A, Kronbach, T, Pangalos, M, Hofgen, N, Brandon, N.J. | | Deposit date: | 2011-06-29 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Highly Potent, Selective, and Orally Active Phosphodiesterase 10A Inhibitors.
J.Med.Chem., 54, 2011
|
|
4J2H
 
 | | Crystal structure of a putative short-chain alcohol dehydrogenase from Sinorhizobium meliloti 1021 (Target NYSGRC-011708) | | Descriptor: | 1,2-ETHANEDIOL, PENTAETHYLENE GLYCOL, SODIUM ION, ... | | Authors: | Sampathkumar, P, Gizzi, A, Ahmed, M, Banu, N, Bhosle, R, Bonanno, J, Chamala, S, Chowdhury, S, Fiser, A, Glenn, A.S, Hammonds, J, Hillerich, B, Khafizov, K, Lafleur, J, Love, J.D, Stead, M, Seidel, R, Toro, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-04 | | Release date: | 2013-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a putative short-chain alcohol dehydrogenase from Sinorhizobium meliloti 1021 (Target NYSGRC-011708)
to be published
|
|
5V7N
 
 | | Crystal structure of NADPH-dependent glyoxylate/hydroxypyruvate reductase SMc04462 (SmGhrB) from Sinorhizobium meliloti in complex with NADP and 2-Keto-D-gluconic acid | | Descriptor: | 2-keto-D-gluconic acid, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shabalin, I.G, Handing, K.B, Miks, C.D, Kutner, J, Matelska, D, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2017-03-20 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
Biochemistry, 57, 2018
|
|
5V7G
 
 | | Crystal structure of NADPH-dependent glyoxylate/hydroxypyruvate reductase SMc04462 (SmGhrB) from Sinorhizobium meliloti in complex with NADPH and oxalate | | Descriptor: | CHLORIDE ION, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Shabalin, I.G, Mason, D.V, Handing, K.B, Kutner, J, Matelska, D, Cooper, D.R, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2017-03-20 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
Biochemistry, 57, 2018
|
|
5TS8
 
 | | Z. MAYS CK2 KINASE ALPHA SUBUNIT IN COMPLEX WITH THE ATP-COMPETITIVE INHIBITOR 5,6-DIBROMOBENZOTRIAZOLE | | Descriptor: | 1,2-ETHANEDIOL, 5,6-DIBROMOBENZOTRIAZOLE, ACETATE ION, ... | | Authors: | Winiewska, M, Kucinska, K, Czapinska, H, Piasecka, A, Bochtler, M, Poznanski, J. | | Deposit date: | 2016-10-28 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | STRUCTURAL AND THERMODYNAMIC ANALYSIS OF 5,6-DIBROMOBENZOTRIAZOLE BINDING TO CASEIN KINASE 2 ALPHA
To Be Published
|
|
5UNN
 
 | | Crystal structure of NADPH-dependent glyoxylate/hydroxypyruvate reductase SMc02828 (SmGhrA) from Sinorhizobium meliloti in apo form | | Descriptor: | CHLORIDE ION, GLYCEROL, NADPH-dependent glyoxylate/hydroxypyruvate reductase | | Authors: | Shabalin, I.G, LaRowe, C, Kutner, J, Gasiorowska, O.A, Handing, K.B, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2017-01-31 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
Biochemistry, 57, 2018
|
|
5V6Q
 
 | | Crystal structure of NADPH-dependent glyoxylate/hydroxypyruvate reductase SMc04462 (SmGhrB) from Sinorhizobium meliloti in complex with NADP and malonate | | Descriptor: | MALONIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH-dependent glyoxylate/hydroxypyruvate reductase, ... | | Authors: | Shabalin, I.G, Handing, K.B, Miezaniec, A.P, Gasiorowska, O.A, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2017-03-17 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
Biochemistry, 57, 2018
|
|
4Z0P
 
 | | Crystal structure of NADPH-dependent glyoxylate/hydroxypyruvate reductase SMc02828 (SmGhrA) from Sinorhizobium meliloti in complex with NADPH and oxalate | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Sroka, P, Gasiorowska, O.A, Handing, K.B, Shabalin, I.G, Porebski, P.J, Hillerich, B.S, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-03-26 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
Biochemistry, 57, 2018
|
|
5V72
 
 | | Crystal structure of NADPH-dependent glyoxylate/hydroxypyruvate reductase SMc04462 (SmGhrB) from Sinorhizobium meliloti in complex with citrate | | Descriptor: | CHLORIDE ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Shabalin, I.G, Handing, K.B, Gasiorowska, O.A, Cooper, D.R, Matelska, D, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2017-03-17 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
Biochemistry, 57, 2018
|
|
5UOG
 
 | | Crystal structure of NADPH-dependent glyoxylate/hydroxypyruvate reductase SMc04462 (SmGhrB) from Sinorhizobium meliloti in apo form | | Descriptor: | NADPH-dependent glyoxylate/hydroxypyruvate reductase, SULFATE ION | | Authors: | Shabalin, I.G, Handing, K.B, Gasiorowska, O.A, Cooper, D.R, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2017-01-31 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
Biochemistry, 57, 2018
|
|
4KTO
 
 | | Crystal Structure Of a Putative Isovaleryl-CoA dehydrogenase (PSI-NYSGRC-012251) from Sinorhizobium meliloti 1021 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FLAVIN-ADENINE DINUCLEOTIDE, isovaleryl-CoA dehydrogenase | | Authors: | Kumar, P.R, Ahmed, M, Attonito, J, Bhosle, R, Bonanno, J, Chamala, S, Chowdhury, S, Glenn, A.S, Hammonds, J, Hillerich, B, Himmel, D, Love, J.D, Seidel, R, Stead, M, Toro, R, Wasserman, S.R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-05-20 | | Release date: | 2013-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.137 Å) | | Cite: | Crystal structure of a Putative Isovaleryl-CoA dehydrogenase from Sinorhizobium meliloti 1021
to be published
|
|
8QCJ
 
 | |
8QCK
 
 | |
4ZQB
 
 | | Crystal structure of NADP-dependent dehydrogenase from Rhodobactersphaeroides in complex with NADP and sulfate | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Kowiel, M, Gasiorowska, O.A, Shabalin, I.G, Handing, K.B, Porebski, P.J, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-05-08 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of NADP-dependent dehydrogenase from Rhodobactersphaeroides in complex with NADP and sulfate
to be published
|
|
4YYC
 
 | | Crystal structure of trimethylamine methyltransferase from Sinorhizobium meliloti in complex with unknown ligand | | Descriptor: | CHLORIDE ION, Putative trimethylamine methyltransferase, UNKNOWN LIGAND | | Authors: | Shabalin, I.G, Porebski, P.J, Gasiorowska, O.A, Handing, K.B, Niedzialkowska, E, Cymborowski, M.T, Cooper, D.R, Stead, M, Hammonds, J, Ahmed, M, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-03-23 | | Release date: | 2015-04-08 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Protein purification and crystallization artifacts: The tale usually not told.
Protein Sci., 25, 2016
|
|
3M7V
 
 | | Crystal structure of phosphopentomutase from streptococcus mutans | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Phosphopentomutase | | Authors: | Fedorov, A.A, Bonanno, J, Fedorov, E.V, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-03-17 | | Release date: | 2010-04-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of phosphopentomutase from streptococcus mutans
To be Published
|
|
3MKV
 
 | | Crystal structure of amidohydrolase eaj56179 | | Descriptor: | CARBONATE ION, GLYCEROL, PUTATIVE AMIDOHYDROLASE, ... | | Authors: | Patskovsky, Y, Bonanno, J, Ozyurt, S, Sauder, J.M, Freeman, J, Wu, B, Smith, D, Bain, K, Rodgers, L, Wasserman, S.R, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-04-15 | | Release date: | 2010-04-28 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional identification and structure determination of two novel prolidases from cog1228 in the amidohydrolase superfamily .
Biochemistry, 49, 2010
|
|
5BP9
 
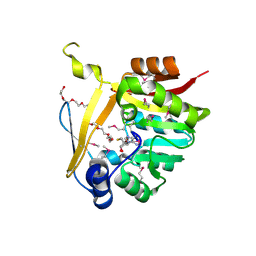 | | Crystal structure of SAM-dependent methyltransferase from Bacteroides fragilis in complex with S-Adenosyl-L-homocysteine | | Descriptor: | 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Putative methyltransferase protein, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Gasiorowska, O.A, Shabalin, I.G, Handing, K.B, Cymborowski, M.T, Mason, D.V, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of SAM-dependent methyltransferase fromBacteroides fragilis in complex with S-Adenosyl-L-homocysteine
to be published
|
|
4KKN
 
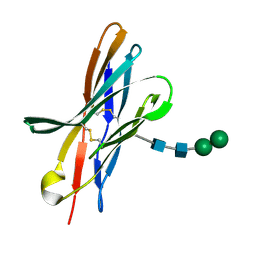 | | Crystal structure of bovine CTLA-4, PSI-NYSGRC-012704 | | Descriptor: | Cytotoxic T-lymphocyte associated protein 4, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kumar, P.R, Ahmed, M, Banu, R, Bhosle, R, Bonanno, J, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chowdhury, S, Fiser, A, Garforth, S.J, Scott Glenn, A, Hammonds, J, Hillerich, B, Khafizov, K, Lafleur, J, Attonito, J, Love, J.D, Patel, H, Patel, R, Seidel, R.D, Smith, B, Stead, M, Toro, R, Casadevall, A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-05-06 | | Release date: | 2013-06-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.253 Å) | | Cite: | Crystal structure of bovine CTLA-4, PSI-NYSGRC-012704
to be published
|
|
