3L8K
 
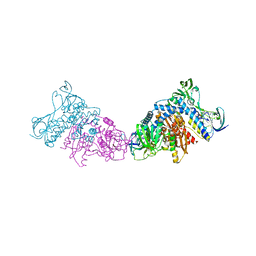 | | Crystal structure of a dihydrolipoyl dehydrogenase from Sulfolobus solfataricus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dihydrolipoyl dehydrogenase, PHOSPHATE ION | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Miller, S, Sampathkumar, P, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-12-31 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a dihydrolipoyl dehydrogenase from Sulfolobus solfataricus
To be Published
|
|
6A7E
 
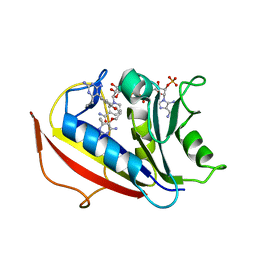 | | Human dihydrofolate reductase complexed with NADPH and BT2 | | Descriptor: | 5-(4-{3-[(2,4-diamino-6-ethylpyrimidin-5-yl)oxy]propoxy}phenyl)-6-ethylpyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Vanichtanankul, J, Tarnchompoo, B, Chitnumsub, P, Kamchonwongpaisan, S, Yuthavong, Y. | | Deposit date: | 2018-07-02 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Hybrid Inhibitors of Malarial Dihydrofolate Reductase with Dual Binding Modes That Can Forestall Resistance.
ACS Med Chem Lett, 9, 2018
|
|
5THG
 
 | |
3KL2
 
 | | Crystal structure of a putative isochorismatase from Streptomyces avermitilis | | Descriptor: | Putative isochorismatase, SULFATE ION | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Chang, S, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-11-06 | | Release date: | 2009-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a putative isochorismatase from Streptomyces avermitilis
To be Published
|
|
6A7C
 
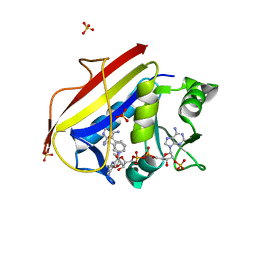 | | Human dihydrofolate reductase complexed with NADPH and BT1 | | Descriptor: | 5-(3-{3-[(2,4-diamino-6-ethylpyrimidin-5-yl)oxy]propoxy}phenyl)-6-ethylpyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Vanichtanankul, J, Tarnchompoo, B, Chitnumsub, P, Kamchonwongpaisan, S, Yuthavong, Y. | | Deposit date: | 2018-07-02 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Hybrid Inhibitors of Malarial Dihydrofolate Reductase with Dual Binding Modes That Can Forestall Resistance.
ACS Med Chem Lett, 9, 2018
|
|
6KP7
 
 | |
3NIW
 
 | | Crystal structure of a haloacid dehalogenase-like hydrolase from Bacteroides thetaiotaomicron | | Descriptor: | GLYCEROL, Haloacid dehalogenase-like hydrolase, MAGNESIUM ION | | Authors: | Bonanno, J.B, Ramagopal, U, Toro, R, Rutter, M, Bain, K.T, Wu, B, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-16 | | Release date: | 2010-06-30 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a haloacid dehalogenase-like hydrolase from Bacteroides thetaiotaomicron
To be Published
|
|
6KP2
 
 | |
6KOT
 
 | |
6KPR
 
 | |
4YIS
 
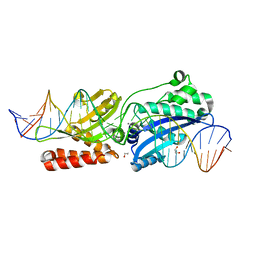 | | Crystal Structure of LAGLIDADG Meganuclease I-CpaMI Bound to Uncleaved DNA | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DNA (28-MER), ... | | Authors: | Hallinan, J.P, Kaiser, B.K, Stoddard, B.L. | | Deposit date: | 2015-03-02 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Indirect DNA Sequence Recognition and Its Impact on Nuclease Cleavage Activity.
Structure, 24, 2016
|
|
4YY2
 
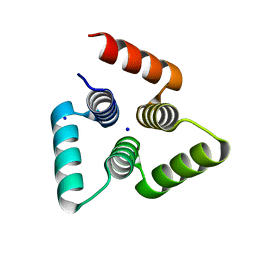 | |
4YIT
 
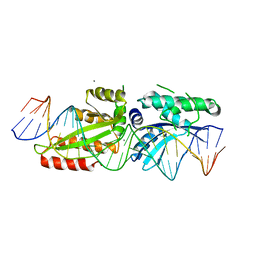 | |
3CYJ
 
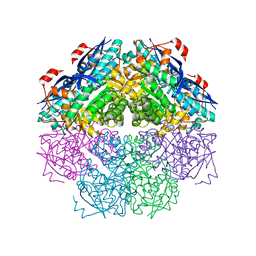 | | Crystal structure of a mandelate racemase/muconate lactonizing enzyme-like protein from Rubrobacter xylanophilus | | Descriptor: | GLYCEROL, Mandelate racemase/muconate lactonizing enzyme-like protein, SODIUM ION | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Zhang, F, Bravo, J, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-25 | | Release date: | 2008-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a mandelate racemase/muconate lactonizing enzyme-like protein from Rubrobacter xylanophilus.
To be Published
|
|
4YHX
 
 | |
4Z1Z
 
 | |
4Z20
 
 | |
3QG2
 
 | | Plasmodium falciparum DHFR-TS qradruple mutant (N51I+C59R+S108N+I164L, V1/S) pyrimethamine complex | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 5-(4-CHLORO-PHENYL)-6-ETHYL-PYRIMIDINE-2,4-DIAMINE, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Vanichtanankul, J, Yuvaniyama, J, Taweechai, S, Chitnumsub, P, Kamchonwongpaisan, S, Yuthavong, Y. | | Deposit date: | 2011-01-24 | | Release date: | 2011-06-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Trypanosomal dihydrofolate reductase reveals natural antifolate resistance
Acs Chem.Biol., 6, 2011
|
|
3GG2
 
 | | Crystal structure of UDP-glucose 6-dehydrogenase from Porphyromonas gingivalis bound to product UDP-glucuronate | | Descriptor: | Sugar dehydrogenase, UDP-glucose/GDP-mannose dehydrogenase family, URIDINE-5'-DIPHOSPHATE-GLUCURONIC ACID | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Sampathkumar, P, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-27 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of UDP-glucose 6-dehydrogenase from Porphyromonas gingivalis bound to product UDP-glucuronate
To be Published
|
|
3C97
 
 | | Crystal structure of the response regulator receiver domain of a signal transduction histidine kinase from Aspergillus oryzae | | Descriptor: | Signal transduction histidine kinase | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-15 | | Release date: | 2008-03-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the response regulator receiver domain of a signal transduction histidine kinase from Aspergillus oryzae.
To be Published
|
|
3H0U
 
 | | Crystal structure of a putative enoyl-CoA hydratase from Streptomyces avermitilis | | Descriptor: | DIMETHYL SULFOXIDE, Putative enoyl-CoA hydratase, SODIUM ION | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Miller, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-10 | | Release date: | 2009-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of a putative enoyl-CoA hydratase from Streptomyces avermitilis
To be Published
|
|
3GKB
 
 | | Crystal structure of a putative enoyl-CoA hydratase from Streptomyces avermitilis | | Descriptor: | GLYCEROL, Putative enoyl-CoA hydratase | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-10 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a putative enoyl-CoA hydratase from Streptomyces avermitilis
To be Published
|
|
3GTZ
 
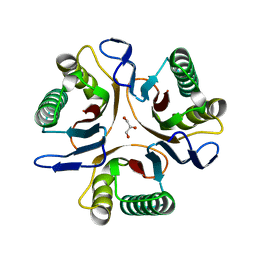 | | Crystal structure of a putative translation initiation inhibitor from Salmonella typhimurium | | Descriptor: | GLYCEROL, Putative translation initiation inhibitor | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Miller, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-28 | | Release date: | 2009-04-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a putative translation initiation inhibitor from Salmonella typhimurium
To be Published
|
|
3E61
 
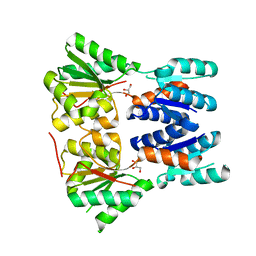 | | Crystal structure of a putative transcriptional repressor of ribose operon from Staphylococcus saprophyticus subsp. saprophyticus | | Descriptor: | GLYCEROL, Putative transcriptional repressor of ribose operon | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-14 | | Release date: | 2008-08-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative transcriptional repressor of ribose operon from Staphylococcus saprophyticus subsp. saprophyticus
To be Published
|
|
3EEZ
 
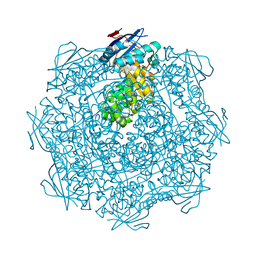 | | Crystal structure of a putative mandelate racemase/muconate lactonizing enzyme from Silicibacter pomeroyi | | Descriptor: | putative Mandelate racemase/muconate lactonizing enzyme | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Hu, S, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-09-07 | | Release date: | 2008-09-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a putative mandelate racemase/muconate lactonizing enzyme from Silicibacter pomeroyi
To be Published
|
|
