2TMV
 
 | |
5H72
 
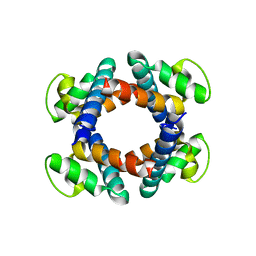 | | Structure of the periplasmic domain of FliP | | Descriptor: | Flagellar biosynthetic protein FliP | | Authors: | Fukumura, T, Kawaguchi, T, Saijo-Hamano, Y, Namba, K, Minamino, T, Imada, K. | | Deposit date: | 2016-11-16 | | Release date: | 2017-08-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Assembly and stoichiometry of the core structure of the bacterial flagellar type III export gate complex
PLoS Biol., 15, 2017
|
|
8YYM
 
 | | Cryo-EM structure of cylindrical fiber of MyD88 TIR | | Descriptor: | Myeloid differentiation primary response protein MyD88 | | Authors: | Kasai, K, Imamura, K, Narita, A, Makino, F, Miyata, T, Kato, T, Namba, K, Onishi, H, Tochio, H. | | Deposit date: | 2024-04-04 | | Release date: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | From Monomers to Oligomers: Structural Mechanism of Receptor-Triggered MyD88 Assembly in Innate Immune Signaling
Biorxiv, 2024
|
|
5H53
 
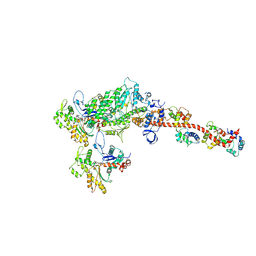 | |
8ROR
 
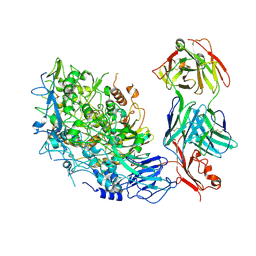 | | Single-particle cryo-EM of Mycoplasma pneumoniae adhesin P1 complexed with the anti-adhesive Fab fragment. | | Descriptor: | Adhesin P1, Heavy Chain Fab, Light Chain Fab | | Authors: | Vizarraga, D, Kawamoto, A, Marcos-Silva, M, Fita, I, Miyata, M, Pinyol, J, Namba, K, Kenri, T. | | Deposit date: | 2024-01-12 | | Release date: | 2025-03-05 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Dynamics of the adhesion complex of the human pathogens Mycoplasma pneumoniae and Mycoplasma genitalium.
Plos Pathog., 21, 2025
|
|
5B0O
 
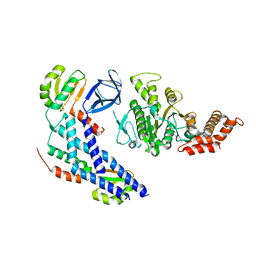 | | Structure of the FliH-FliI complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Flagellar assembly protein FliH, Flagellum-specific ATP synthase | | Authors: | Imada, K, Uchida, Y, Kinoshita, M, Namba, K, Minamino, T. | | Deposit date: | 2015-11-02 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Insight into the flagella type III export revealed by the complex structure of the type III ATPase and its regulator
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1WLG
 
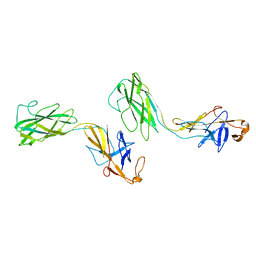 | | Crystal structure of FlgE31, a major fragment of the hook protein | | Descriptor: | Flagellar hook protein flgE | | Authors: | Samatey, F.A, Matsunami, H, Imada, K, Nagashima, S, Shaikh, T.R, Thomas, D.R, DeRosier, D.J, Kitao, A, Namba, K. | | Deposit date: | 2004-06-25 | | Release date: | 2004-11-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the bacterial flagellar hook and implication for the molecular universal joint mechanism.
Nature, 431, 2004
|
|
6LY8
 
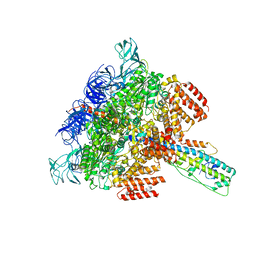 | | V/A-ATPase from Thermus thermophilus, the soluble domain, including V1, d, two EG stalks, and N-terminal domain of a-subunit. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, V-type ATP synthase alpha chain, V-type ATP synthase beta chain, ... | | Authors: | Kishikawa, J, Nakanishi, A, Furuta, A, Kato, T, Namba, K, Tamakoshi, M, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2020-02-13 | | Release date: | 2020-09-09 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanical inhibition of isolated V o from V/A-ATPase for proton conductance.
Elife, 9, 2020
|
|
1VGL
 
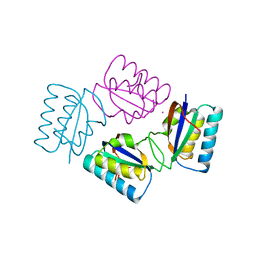 | | Crystal structure of tetrameric KaiB from T.elongatus BP-1 | | Descriptor: | Circadian clock protein kaiB, MERCURY (II) ION | | Authors: | Iwase, R, Imada, K, Hayashi, F, Uzumaki, T, Namba, K, Ishiura, M. | | Deposit date: | 2004-04-27 | | Release date: | 2005-08-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Functionally important substructures of circadian clock protein KaiB in a unique tetramer complex.
J.Biol.Chem., 280, 2005
|
|
3MFP
 
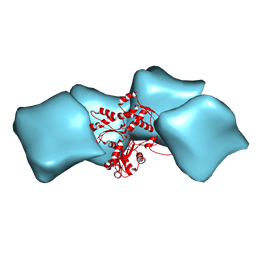 | | Atomic model of F-actin based on a 6.6 angstrom resolution cryoEM map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle | | Authors: | Fujii, T, Iwane, A.H, Yanagida, T, Namba, K. | | Deposit date: | 2010-04-03 | | Release date: | 2010-09-29 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Direct visualization of secondary structures of F-actin by electron cryomicroscopy
Nature, 467, 2010
|
|
1UCU
 
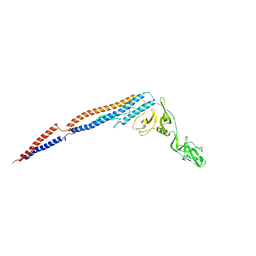 | |
9IWQ
 
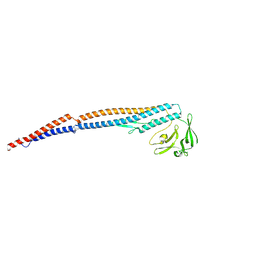 | | Salmonella enterica serovar Typhimurium FliC(G426A)delta(204-292) forming the L-type straight filament | | Descriptor: | Flagellin | | Authors: | Waraich, K, Makino, F, Miyata, T, Kinoshita, M, Minamino, T, Namba, K. | | Deposit date: | 2024-07-25 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.08 Å) | | Cite: | Salmonella enterica serovar Typhimurium FliC(G426A)delta(204-292) forming the L-type straight filament
To Be Published
|
|
5WRH
 
 | |
1GTT
 
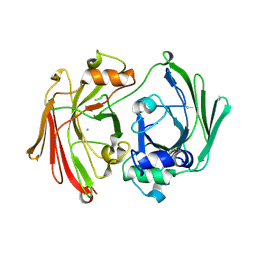 | | CRYSTAL STRUCTURE OF HPCE | | Descriptor: | 4-HYDROXYPHENYLACETATE DEGRADATION BIFUNCTIONAL ISOMERASE/DECARBOXYLASE, CALCIUM ION | | Authors: | Tame, J.R.H, Namba, K, Dodson, E.J, Roper, D.I. | | Deposit date: | 2002-01-18 | | Release date: | 2002-03-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of Hpce, a Bifunctional Decarboxylase/Isomerase with a Multifunctional Fold.
Biochemistry, 41, 2002
|
|
4D3E
 
 | | Tetramer of IpaD, modified from 2J0O, fitted into negative stain electron microscopy reconstruction of the wild type tip complex from the type III secretion system of Shigella flexneri | | Descriptor: | INVASIN IPAD | | Authors: | Cheung, M, Shen, D.-K, Makino, F, Kato, T, Roehrich, D, Martinez-Argudo, I, Walker, M.L, Murillo, I, Liu, X, Pain, M, Brown, J, Frazer, G, Mantell, J, Mina, P, Todd, T, Sessions, R.B, Namba, K, Blocker, A.J. | | Deposit date: | 2014-10-21 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (24 Å) | | Cite: | Three-Dimensional Electron Microscopy Reconstruction and Cysteine-Mediated Crosslinking Provide a Model of the T3Ss Needle Tip Complex.
Mol.Microbiol., 95, 2015
|
|
5B3D
 
 | | Structure of a flagellar type III secretion chaperone, FlgN | | Descriptor: | Flagella synthesis protein FlgN | | Authors: | Nakanishi, Y, Kinoshita, M, Namba, K, Minamino, T, Imada, K. | | Deposit date: | 2016-02-15 | | Release date: | 2016-06-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rearrangements of alpha-helical structures of FlgN chaperone control the binding affinity for its cognate substrates during flagellar type III export
Mol.Microbiol., 101, 2016
|
|
1A05
 
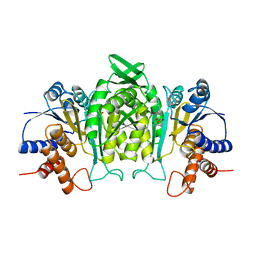 | | CRYSTAL STRUCTURE OF THE COMPLEX OF 3-ISOPROPYLMALATE DEHYDROGENASE FROM THIOBACILLUS FERROOXIDANS WITH 3-ISOPROPYLMALATE | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE, 3-ISOPROPYLMALIC ACID, MAGNESIUM ION | | Authors: | Imada, K, Inagaki, K, Matsunami, H, Kawaguchi, H, Tanaka, H, Tanaka, N, Namba, K. | | Deposit date: | 1997-12-09 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of 3-isopropylmalate dehydrogenase in complex with 3-isopropylmalate at 2.0 A resolution: the role of Glu88 in the unique substrate-recognition mechanism.
Structure, 6, 1998
|
|
3J6P
 
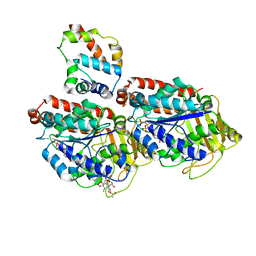 | | Pseudo-atomic model of dynein microtubule binding domain-tubulin complex based on a cryoEM map | | Descriptor: | Dynein heavy chain, cytoplasmic, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Uchimura, S, Fujii, T, Takazaki, H, Ayukawa, R, Nishikawa, Y, Minoura, I, Hachikubo, Y, Kurisu, G, Sutoh, K, Kon, T, Namba, K, Muto, E. | | Deposit date: | 2014-03-20 | | Release date: | 2014-12-31 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | A flipped ion pair at the dynein-microtubule interface is critical for dynein motility and ATPase activation
J.Cell Biol., 208, 2015
|
|
6LU1
 
 | | Cyanobacterial PSI Monomer from T. elongatus by Single Particle CRYO-EM at 3.2 A Resolution | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Kurisu, G, Coruh, O, Tanaka, H, Gerle, C, Kawamoto, A, Kato, T, Namba, K, Nowaczyk, M.M, Rogner, M, Misumi, Y, Frank, A, Eithar, E.M. | | Deposit date: | 2020-01-24 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of a functional monomeric Photosystem I from Thermosynechococcus elongatus reveals red chlorophyll cluster.
Commun Biol, 4, 2021
|
|
3TEE
 
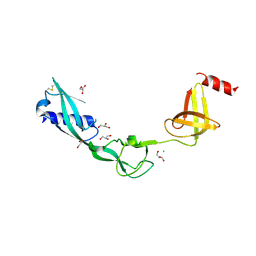 | | Crystal Structure of Salmonella FlgA in open form | | Descriptor: | CHLORIDE ION, Flagella basal body P-ring formation protein flgA, GLYCEROL | | Authors: | Matsunami, H, Samatey, F.A, Namba, K. | | Deposit date: | 2011-08-12 | | Release date: | 2012-08-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural flexibility of the periplasmic protein, FlgA, regulates flagellar P-ring assembly in Salmonella enterica
Sci Rep, 6, 2016
|
|
7VQ0
 
 | | Cryo-EM structure of the SARS-CoV-2 spike protein (2-up RBD) bound to neutralizing nanobodies P86 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Maeda, R, Fujita, J, Konishi, Y, Kazuma, Y, Yamazaki, H, Anzai, I, Yamaguchi, K, Kasai, K, Nagata, K, Yamaoka, Y, Miyakawa, K, Ryo, A, Shirakawa, K, Makino, F, Matsuura, Y, Inoue, T, Imura, A, Namba, K, Takaori-Kondo, A. | | Deposit date: | 2021-10-18 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | A panel of nanobodies recognizing conserved hidden clefts of all SARS-CoV-2 spike variants including Omicron.
Commun Biol, 5, 2022
|
|
8HLB
 
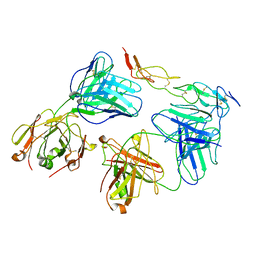 | | Cryo-EM structure of biparatopic antibody Bp109-92 in complex with TNFR2 | | Descriptor: | TR109 heavy chain, TR109 light chain, TR92 heavy chain, ... | | Authors: | Akiba, H, Fujita, J, Ise, T, Nishiyama, K, Miyata, T, Kato, T, Namba, K, Ohno, H, Kamada, H, Nagata, S, Tsumoto, K. | | Deposit date: | 2022-11-29 | | Release date: | 2023-10-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Development of a 1:1-binding biparatopic anti-TNFR2 antagonist by reducing signaling activity through epitope selection.
Commun Biol, 6, 2023
|
|
8JEK
 
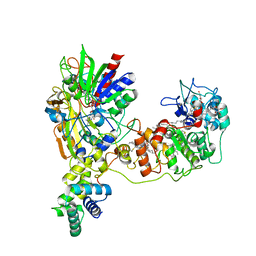 | | Cryo-EM Structure of K-ferricyanide Oxidized Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Fructose dehydrogenase cytochrome subunit, ... | | Authors: | Suzuki, Y, Miyata, T, Makino, F, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2023-05-16 | | Release date: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Essential Insight of Direct Electron Transfer-Type Bioelectrocatalysis by Membrane-Bound d-Fructose Dehydrogenase with Structural Bioelectrochemistry.
Acs Catalysis, 13, 2023
|
|
8JEJ
 
 | | Cryo-EM Structure of Na-dithionite Reduced Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Fructose dehydrogenase cytochrome subunit, ... | | Authors: | Suzuki, Y, Miyata, T, Makino, F, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2023-05-16 | | Release date: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Essential Insight of Direct Electron Transfer-Type Bioelectrocatalysis by Membrane-Bound d-Fructose Dehydrogenase with Structural Bioelectrochemistry.
Acs Catalysis, 13, 2023
|
|
1IO1
 
 | | CRYSTAL STRUCTURE OF F41 FRAGMENT OF FLAGELLIN | | Descriptor: | PHASE 1 FLAGELLIN | | Authors: | Samatey, F.A, Imada, K, Nagashima, S, Vondervisz, F, Kumasaka, T, Yamamoto, M, Namba, K. | | Deposit date: | 2000-12-28 | | Release date: | 2001-04-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the bacterial flagellar protofilament and implications for a switch for supercoiling
Nature, 410, 2001
|
|
