3DL8
 
 | | Structure of the complex of aquifex aeolicus SecYEG and bacillus subtilis SecA | | Descriptor: | Preprotein translocase subunit secY, Protein translocase subunit secA, Protein-export membrane protein secG, ... | | Authors: | Nam, Y, Zimmer, J, Rapoport, T.A. | | Deposit date: | 2008-06-26 | | Release date: | 2008-12-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (7.5 Å) | | Cite: | Structure of a complex of the ATPase SecA and the protein-translocation channel.
Nature, 455, 2008
|
|
5UDZ
 
 | |
3TS0
 
 | | Mouse Lin28A in complex with let-7f-1 microRNA pre-element | | Descriptor: | Protein lin-28 homolog A, RNA (5'-R(*GP*GP*GP*GP*UP*AP*GP*UP*GP*AP*UP*UP*UP*UP*AP*CP*CP*CP*UP*GP*GP*AP*G)-3'), ZINC ION | | Authors: | Nam, Y, Sliz, P. | | Deposit date: | 2011-09-11 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.763 Å) | | Cite: | Molecular Basis for Interaction of let-7 MicroRNAs with Lin28.
Cell(Cambridge,Mass.), 147, 2011
|
|
3TRZ
 
 | | Mouse Lin28A in complex with let-7d microRNA pre-element | | Descriptor: | Protein lin-28 homolog A, RNA (5'-R(*GP*GP*GP*CP*AP*GP*GP*GP*AP*UP*UP*UP*UP*GP*CP*CP*CP*GP*GP*AP*G)-3'), ZINC ION | | Authors: | Nam, Y, Sliz, P. | | Deposit date: | 2011-09-11 | | Release date: | 2011-11-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular Basis for Interaction of let-7 MicroRNAs with Lin28.
Cell(Cambridge,Mass.), 147, 2011
|
|
3TS2
 
 | | Mouse Lin28A in complex with let-7g microRNA pre-element | | Descriptor: | Protein lin-28 homolog A, RNA (5'-R(*GP*GP*GP*GP*UP*CP*UP*AP*UP*GP*AP*UP*AP*CP*CP*AP*CP*CP*CP*CP*GP*GP*AP*G)-3'), ZINC ION | | Authors: | Nam, Y, Sliz, P. | | Deposit date: | 2011-09-11 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Molecular Basis for Interaction of let-7 MicroRNAs with Lin28.
Cell(Cambridge,Mass.), 147, 2011
|
|
3V79
 
 | | Structure of human Notch1 transcription complex including CSL, RAM, ANK, and MAML-1 on HES-1 promoter DNA sequence | | Descriptor: | DNA 5'-D(*GP*TP*TP*AP*CP*TP*GP*TP*GP*GP*GP*AP*AP*AP*GP*AP*AP*A)-3', DNA 5'-D(*TP*TP*TP*CP*TP*TP*TP*CP*CP*CP*AP*CP*AP*GP*TP*AP*AP*C)-3', Mastermind-like protein 1, ... | | Authors: | Nam, Y, Sliz, P, Blacklow, S. | | Deposit date: | 2011-12-20 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | Conformational Locking upon Cooperative Assembly of Notch Transcription Complexes.
Structure, 20, 2012
|
|
2F8Y
 
 | | Crystal structure of human Notch1 ankyrin repeats to 1.55A resolution. | | Descriptor: | Notch homolog 1, translocation-associated (Drosophila), SULFATE ION | | Authors: | Nam, Y, Sliz, P, Blacklow, S.C. | | Deposit date: | 2005-12-04 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes.
Cell(Cambridge,Mass.), 124, 2006
|
|
2F8X
 
 | | Crystal structure of activated Notch, CSL and MAML on HES-1 promoter DNA sequence | | Descriptor: | 5'-D(*GP*TP*TP*AP*CP*TP*GP*TP*GP*GP*GP*AP*AP*AP*GP*AP*AP*A)-3', 5'-D(*TP*TP*TP*CP*TP*TP*TP*CP*CP*CP*AP*CP*AP*GP*TP*AP*AP*C)-3', Mastermind-like protein 1, ... | | Authors: | Nam, Y, Sliz, P, Blacklow, S.C. | | Deposit date: | 2005-12-04 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes.
Cell(Cambridge,Mass.), 124, 2006
|
|
5AWJ
 
 | | Crystal structure of VDR-LBD/partial agonist complex: 22S-hexyl analogue | | Descriptor: | (1R,3R)-5-[(2E)-2-[(1R,3aS,7aR)-1-[(2R,3S)-3-(2-hydroxyethyl)nonan-2-yl]-7a-methyl-2,3,3a,5,6,7-hexahydro-1H-inden-4-ylidene]ethylidene]-2-methylidene-cyclohexane-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor,Vitamin D3 receptor | | Authors: | Anami, Y, Itoh, T, Inaba, Y, Nakabayashi, M, Ikura, T, Ito, N, Yamamoto, K. | | Deposit date: | 2015-07-04 | | Release date: | 2015-11-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Fine tuning of agonistic/antagonistic activity for vitamin D receptor by 22-alkyl chain length of ligands: 22S-Hexyl compound unexpectedly restored agonistic activity.
Bioorg.Med.Chem., 23, 2015
|
|
5AWK
 
 | | Crystal structure of VDR-LBD/partial agonist complex: 22S-ethyl analogue | | Descriptor: | (1R,3R)-5-[(2E)-2-[(1R,3aS,7aR)-1-[(2R,3S)-3-ethyl-5-oxidanyl-pentan-2-yl]-7a-methyl-2,3,3a,5,6,7-hexahydro-1H-inden-4-ylidene]ethylidene]-2-methylidene-cyclohexane-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor,Vitamin D3 receptor | | Authors: | Anami, Y, Itoh, T, Yamamoto, K. | | Deposit date: | 2015-07-04 | | Release date: | 2015-11-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Fine tuning of agonistic/antagonistic activity for vitamin D receptor by 22-alkyl chain length of ligands: 22S-Hexyl compound unexpectedly restored agonistic activity.
Bioorg.Med.Chem., 23, 2015
|
|
5B41
 
 | | Crystal structure of VDR-LBD complexed with 2-methylidene-19-nor-1a,25-dihydroxyvitamin D3 | | Descriptor: | (1R,3R)-5-(2-((1R,3aS,7aR,E)-1-((R)-6-hydroxy-6-methylheptan-2-yl)-7a-methyloctahydro-4H-inden-4-ylidene)ethylidene)-2- methylenecyclohexane-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Anami, Y, Itoh, T, Yamamoto, K. | | Deposit date: | 2016-03-23 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Apo- and Antagonist-Binding Structures of Vitamin D Receptor Ligand-Binding Domain Revealed by Hybrid Approach Combining Small-Angle X-ray Scattering and Molecular Dynamics
J.Med.Chem., 59, 2016
|
|
3WT5
 
 | | A mixed population of antagonist and agonist binding conformers in a single crystal explains partial agonism against vitamin D receptor: Active vitamin D analogues with 22R-alkyl group | | Descriptor: | (1R,3R,7E,17beta)-17-[(2R,3R)-3-butyl-6-hydroxy-6-methylheptan-2-yl]-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Anami, Y, Itoh, T, Yamamoto, K. | | Deposit date: | 2014-04-08 | | Release date: | 2014-06-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Mixed Population of Antagonist and Agonist Binding Conformers in a Single Crystal Explains Partial Agonism against Vitamin D Receptor: Active Vitamin D Analogues with 22R-Alkyl Group.
J.Med.Chem., 57, 2014
|
|
3WT7
 
 | | Crystal structure of VDR-LBD complexed with 22R-Butyl-2-methylidene-26,27-dimethyl-19,24-dinor-1 ,25-dihydroxyvitamin D3 | | Descriptor: | (1R,3R,7E,17beta)-17-[(2R,3R)-3-butyl-5-ethyl-5-hydroxyheptan-2-yl]-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Anami, Y, Itoh, T, Yamamoto, K. | | Deposit date: | 2014-04-08 | | Release date: | 2014-06-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Mixed Population of Antagonist and Agonist Binding Conformers in a Single Crystal Explains Partial Agonism against Vitamin D Receptor: Active Vitamin D Analogues with 22R-Alkyl Group.
J.Med.Chem., 57, 2014
|
|
3WT6
 
 | | A mixed population of antagonist and agonist binding conformers in a single crystal explains partial agonism against vitamin D receptor: Active vitamin D analogues with 22R-alkyl group | | Descriptor: | (1R,3R,7E,17beta)-17-[(2R,3R)-3-butyl-6-hydroxy-6-methylheptan-2-yl]-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Anami, Y, Itoh, T, Yamamoto, K. | | Deposit date: | 2014-04-08 | | Release date: | 2014-06-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Mixed Population of Antagonist and Agonist Binding Conformers in a Single Crystal Explains Partial Agonism against Vitamin D Receptor: Active Vitamin D Analogues with 22R-Alkyl Group.
J.Med.Chem., 57, 2014
|
|
6DU4
 
 | | Crystal structure of hMettl16 catalytic domain in complex with MAT2A 3'UTR hairpin 1 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, U6 small nuclear RNA (adenine-(43)-N(6))-methyltransferase, ... | | Authors: | Doxtader, K, Wang, P, Nam, Y. | | Deposit date: | 2018-06-19 | | Release date: | 2018-09-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Regulation of METTL16, an S-Adenosylmethionine Homeostasis Factor.
Mol. Cell, 71, 2018
|
|
6V5C
 
 | | Human Drosha and DGCR8 in complex with Primary MicroRNA (MP/RNA complex) - partially docked state | | Descriptor: | Microprocessor complex subunit DGCR8, Pri-miR-16-2 (66-MER), Ribonuclease 3 | | Authors: | Partin, A, Zhang, K, Jeong, B, Herrell, E, Li, S, Chiu, W, Nam, Y. | | Deposit date: | 2019-12-04 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM Structures of Human Drosha and DGCR8 in Complex with Primary MicroRNA.
Mol.Cell, 78, 2020
|
|
6UV4
 
 | | Crystal structure of the core domain of RNA helicase DDX17 with RNA pri-18a-oligo1 | | Descriptor: | 18a_oligo1, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Ngo, T.D, Partin, A.C, Nam, Y. | | Deposit date: | 2019-11-01 | | Release date: | 2020-01-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | RNA Specificity and Autoregulation of DDX17, a Modulator of MicroRNA Biogenesis.
Cell Rep, 29, 2019
|
|
6UV3
 
 | | Crystal structure of the core domain of RNA helicase DDX17 with RNA pri-125a-oligo2 | | Descriptor: | 125a-oligo2, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Ngo, T.D, Partin, A.C, Nam, Y. | | Deposit date: | 2019-11-01 | | Release date: | 2020-01-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | RNA Specificity and Autoregulation of DDX17, a Modulator of MicroRNA Biogenesis.
Cell Rep, 29, 2019
|
|
6UV0
 
 | |
6UV1
 
 | | Crystal structure of RNA helicase DDX17 in complex of rU10 RNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Ngo, T.D, Partin, A.C, Nam, Y. | | Deposit date: | 2019-11-01 | | Release date: | 2020-01-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.307 Å) | | Cite: | RNA Specificity and Autoregulation of DDX17, a Modulator of MicroRNA Biogenesis.
Cell Rep, 29, 2019
|
|
6UV2
 
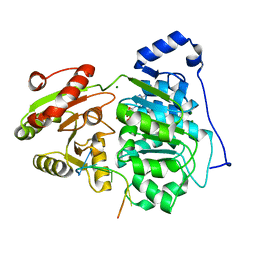 | | Crystal structure of the core domain of RNA helicase DDX17 with RNA pri-125a-oligo1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Ngo, T.D, Partin, A.C, Nam, Y. | | Deposit date: | 2019-11-01 | | Release date: | 2020-01-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.894 Å) | | Cite: | RNA Specificity and Autoregulation of DDX17, a Modulator of MicroRNA Biogenesis.
Cell Rep, 29, 2019
|
|
6V5B
 
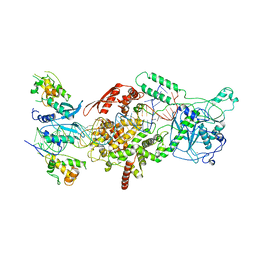 | | Human Drosha and DGCR8 in complex with Primary MicroRNA (MP/RNA complex) - Active state | | Descriptor: | CALCIUM ION, Microprocessor complex subunit DGCR8, Pri-miR-16-2 (78-MER), ... | | Authors: | Partin, A, Zhang, K, Jeong, B, Herrell, E, Li, S, Chiu, W, Nam, Y. | | Deposit date: | 2019-12-04 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM Structures of Human Drosha and DGCR8 in Complex with Primary MicroRNA.
Mol.Cell, 78, 2020
|
|
7RX6
 
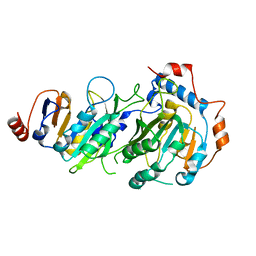 | |
7RX8
 
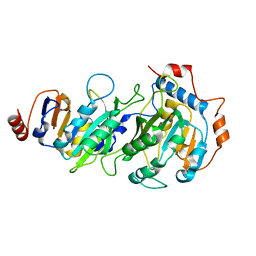 | |
5K7U
 
 | |
