1X26
 
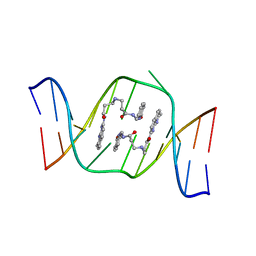 | | Solution structure of the AA-mismatch DNA complexed with naphthyridine-azaquinolone | | Descriptor: | 5'-D(*CP*AP*TP*TP*CP*AP*GP*TP*TP*AP*G)-3', 5'-D(*CP*TP*AP*AP*CP*AP*GP*AP*AP*TP*G)-3', N~3~-{3-[(7-METHYL-1,8-NAPHTHYRIDIN-2-YL)AMINO]-3-OXOPROPYL}-N~1~-[(7-OXO-7,8-DIHYDRO-1,8-NAPHTHYRIDIN-2-YL)METHYL]-BET A-ALANINAMIDE | | Authors: | Nakatani, K, Hagihara, S, Goto, Y, Kobori, A, Hagihara, M, Hayashi, G, Kyo, M, Nomura, M, Mishima, M, Kojima, C. | | Deposit date: | 2005-04-20 | | Release date: | 2006-04-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Small-molecule ligand induces nucleotide flipping in (CAG)n trinucleotide repeats
Nat.Chem.Biol., 1, 2005
|
|
7YVW
 
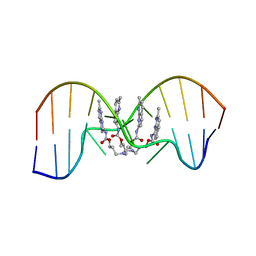 | | NMR determination of the 2:1 binding motif structure involving cytosine flipping out for the recognition of the CGG/CGG triad DNA | | Descriptor: | 3-[3-[(7-methyl-1,8-naphthyridin-2-yl)carbamoyloxy]propylamino]propyl ~{N}-(7-methyl-1,8-naphthyridin-2-yl)carbamate, DNA (5'-D(*CP*AP*TP*TP*CP*GP*GP*TP*TP*AP*G)-3'), DNA (5'-D(*CP*TP*AP*AP*CP*GP*GP*AP*AP*TP*G)-3') | | Authors: | Furuita, K, Yamada, T, Sakurabayashi, S, Nomura, M, Kojima, C, Nakatani, K. | | Deposit date: | 2022-08-19 | | Release date: | 2023-06-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR determination of the 2:1 binding complex of naphthyridine carbamate dimer (NCD) and CGG/CGG triad in double-stranded DNA.
Nucleic Acids Res., 50, 2022
|
|
7CPK
 
 | | Xylanase R from Bacillus sp. TAR-1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Endo-1,4-beta-xylanase A, ... | | Authors: | Kuwata, K, Suzuki, M, Takita, T, Nakatani, K, Li, T, Katano, Y, Kojima, K, Mizutani, K, Mikami, B, Yatsunami, R, Nakamura, S, Yasukawa, K. | | Deposit date: | 2020-08-07 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insight into the mechanism of thermostabilization of GH10 xylanase from Bacillus sp. strain TAR-1 by the mutation of S92 to E.
Biosci.Biotechnol.Biochem., 85, 2021
|
|
7CPL
 
 | | Xylanase R from Bacillus sp. TAR-1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Endo-1,4-beta-xylanase A, ... | | Authors: | Kuwata, K, Suzuki, M, Takita, T, Nakatani, K, Li, T, Katano, Y, Kojima, K, Mizutani, K, Mikami, B, Yatsunami, R, Nakamura, S, Yasukawa, K. | | Deposit date: | 2020-08-07 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Insight into the mechanism of thermostabilization of GH10 xylanase from Bacillus sp. strain TAR-1 by the mutation of S92 to E.
Biosci.Biotechnol.Biochem., 85, 2021
|
|
8ZD8
 
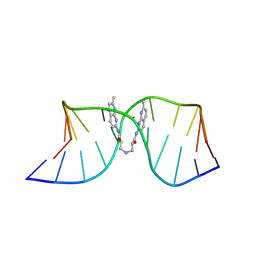 | | NMR structure of the (CGG-dsDNA:ND=) 1:1 complex | | Descriptor: | DNA (5'-D(*CP*AP*TP*TP*CP*GP*GP*TP*TP*AP*G)-3'), DNA (5'-D(*CP*TP*AP*AP*CP*GP*GP*AP*AP*TP*G)-3'), ~{N}-(7-methyl-1,8-naphthyridin-2-yl)-3-[[3-[(7-methyl-1,8-naphthyridin-2-yl)amino]-3-oxidanylidene-propyl]amino]propanamide | | Authors: | Sakurabayashi, S, Furuita, K, Yamada, T, Nomura, M, Nakatani, K, Kojima, C. | | Deposit date: | 2024-05-01 | | Release date: | 2025-04-30 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the (CGG-dsDNA:ND=) 1:1 complex
To Be Published
|
|
8ZD7
 
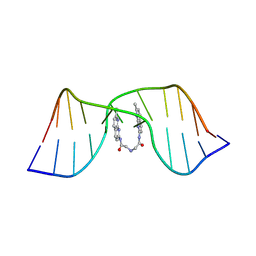 | | NMR structure of the CGG-dsDNA-sND complex | | Descriptor: | DNA (5'-D(*CP*AP*TP*TP*CP*GP*GP*TP*TP*AP*G)-3'), DNA (5'-D(*CP*TP*AP*AP*CP*GP*GP*AP*AP*TP*G)-3'), ~{N}-(7-methyl-1,8-naphthyridin-2-yl)-2-[[2-[(7-methyl-1,8-naphthyridin-2-yl)amino]-2-oxidanylidene-ethyl]amino]ethanamide | | Authors: | Sakurabayashi, S, Furuita, K, Yamada, T, Nomura, M, Nakatani, K, Kojima, C. | | Deposit date: | 2024-05-01 | | Release date: | 2025-04-30 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the CGG-dsDNA-sND complex
To Be Published
|
|
8ZD2
 
 | | NMR structure of the (CGG-dsDNA:ND=) 1:2 complex | | Descriptor: | DNA (5'-D(*CP*AP*TP*TP*CP*GP*GP*TP*TP*AP*G)-3'), DNA (5'-D(*CP*TP*AP*AP*CP*GP*GP*AP*AP*TP*G)-3'), ~{N}-(7-methyl-1,8-naphthyridin-2-yl)-3-[[3-[(7-methyl-1,8-naphthyridin-2-yl)amino]-3-oxidanylidene-propyl]amino]propanamide | | Authors: | Sakurabayashi, S, Furuita, K, Yamada, T, Nomura, M, Nakatani, K, Kojima, C. | | Deposit date: | 2024-05-01 | | Release date: | 2025-04-30 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the (CGG-dsDNA:ND=) 1:2 complex
To Be Published
|
|
2ELG
 
 | | The rare crystallographic structure of d(CGCGCG)2: The natural spermidine molecule bound to the minor groove of left-handed Z-DNA d(CGCGCG)2 at 10 degree celsius | | Descriptor: | DNA (5'-D(*DCP*DGP*DCP*DGP*DCP*DG)-3'), MAGNESIUM ION, SODIUM ION, ... | | Authors: | Ohishi, H, Tozuka, Y, Zhou, D.Y, Ishida, T, Nakatani, K. | | Deposit date: | 2007-03-27 | | Release date: | 2008-04-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The rare crystallographic structure of d(CGCGCG)(2): The natural spermidine molecule bound to the minor groove of left-handed Z-DNA d(CGCGCG)(2) at 10 degrees C
Biochem.Biophys.Res.Commun., 358, 2007
|
|
6KJL
 
 | | Xylanase J from Bacillus sp. strain 41M-1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Manami, S, Teisuke, T, Nakatani, K, Katano, K, Kojima, K, Saka, N, Mikami, B, Yatsunami, R, Nakamura, S, Yasukawa, K. | | Deposit date: | 2019-07-22 | | Release date: | 2019-09-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Increase in the thermostability of GH11 xylanase XynJ from Bacillus sp. strain 41M-1 using site saturation mutagenesis.
Enzyme.Microb.Technol., 130, 2019
|
|
6IZP
 
 | | Solution structure of the complex of naphthyridine carbamate dimer and an RNA with UGGAA-UGGAA pentad | | Descriptor: | 3-[3-[(7-methyl-1,8-naphthyridin-2-yl)carbamoyloxy]propylamino]propyl ~{N}-(7-methyl-1,8-naphthyridin-2-yl)carbamate, RNA (29-MER) | | Authors: | Nagano, K, Shibata, T, Nakatani, K, Kawai, G. | | Deposit date: | 2018-12-20 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Small molecule targeting r(UGGAA)n disrupts RNA foci and alleviates disease phenotype in Drosophila model
Nat Commun, 12, 2021
|
|
6KKA
 
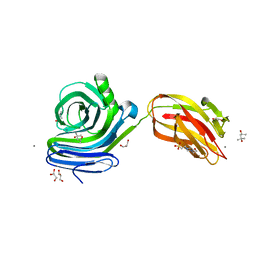 | | Xylanase J mutant from Bacillus sp. 41M-1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Suzuki, M, Takita, T, Nakatani, K. | | Deposit date: | 2019-07-24 | | Release date: | 2019-09-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Increase in the thermostability of GH11 xylanase XynJ from Bacillus sp. strain 41M-1 using site saturation mutagenesis.
Enzyme.Microb.Technol., 130, 2019
|
|
6QIS
 
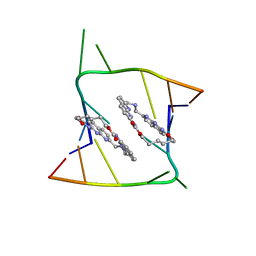 | | Crystal structure of CAG repeats with synthetic CMBL3a compound (model II) | | Descriptor: | CMBL3a, RNA (5'-R(*GP*CP*AP*GP*CP*AP*GP*C)-3'), SULFATE ION | | Authors: | Kiliszek, A, Blaszczyk, L, Rypniewski, W, Nakatani, K. | | Deposit date: | 2019-01-21 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural insights into synthetic ligands targeting A-A pairs in disease-related CAG RNA repeats.
Nucleic Acids Res., 47, 2019
|
|
6QIV
 
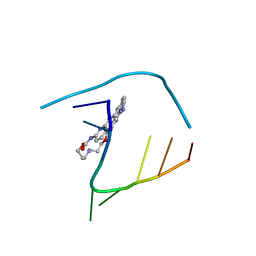 | | Crystal structure of seleno-derivative CAG repeats with synthetic CMBL4 compound | | Descriptor: | CMBL4, RNA (5'-R(*GP*CP*AP*GP*CP*AP*GP*C)-3') | | Authors: | Kiliszek, A, Blaszczyk, L, Rypniewski, W, Nakatani, K. | | Deposit date: | 2019-01-21 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural insights into synthetic ligands targeting A-A pairs in disease-related CAG RNA repeats.
Nucleic Acids Res., 47, 2019
|
|
6QIR
 
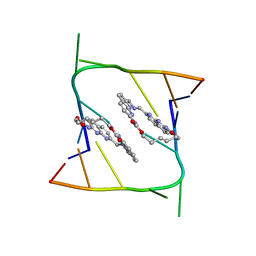 | | Crystal structure of CAG repeats with synthetic CMBL3a compound (model I) | | Descriptor: | CMBL3a, RNA (5'-R(*GP*CP*AP*GP*CP*AP*GP*C)-3') | | Authors: | Kiliszek, A, Blaszczyk, L, Rypniewski, W, Nakatani, K. | | Deposit date: | 2019-01-21 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.531 Å) | | Cite: | Structural insights into synthetic ligands targeting A-A pairs in disease-related CAG RNA repeats.
Nucleic Acids Res., 47, 2019
|
|
6QIQ
 
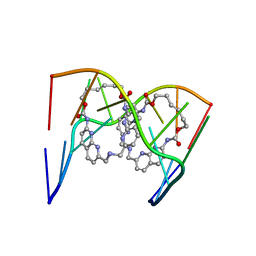 | | Crystal structure of seleno-derivative CAG repeats with synthetic CMBL3a compound | | Descriptor: | CMBL3a, RNA (5'-R(*GP*CP*AP*G)-D(P*(CSL))-R(P*AP*GP*C)-3') | | Authors: | Kiliszek, A, Blaszczyk, L, Rypniewski, W, Micura, R, Nakatani, K. | | Deposit date: | 2019-01-21 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.519 Å) | | Cite: | Structural insights into synthetic ligands targeting A-A pairs in disease-related CAG RNA repeats.
Nucleic Acids Res., 47, 2019
|
|
6QIT
 
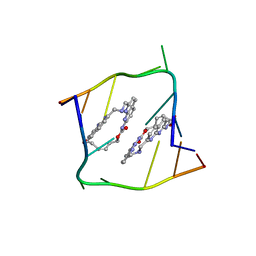 | | Crystal structure of CAG repeats with synthetic CMBL3b compound | | Descriptor: | CMBL3a, RNA (5'-R(*GP*CP*AP*GP*CP*AP*GP*C)-3') | | Authors: | Kiliszek, A, Blaszczyk, L, Rypniewski, W, Nakatani, K. | | Deposit date: | 2019-01-21 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Structural insights into synthetic ligands targeting A-A pairs in disease-related CAG RNA repeats.
Nucleic Acids Res., 47, 2019
|
|
