2N90
 
 | |
2MIC
 
 | |
2LOH
 
 | |
2MZF
 
 | | Purotoxin-2 NMR structure in water | | Descriptor: | Purotoxin-2 | | Authors: | Nadezhdin, K, Vassilevski, A, Oparin, P, Grishin, E, Arseniev, A. | | Deposit date: | 2015-02-12 | | Release date: | 2016-04-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of purotoxin-2 from wolf spider: modular design and membrane-assisted mode of action in arachnid toxins.
Biochem. J., 473, 2016
|
|
2N85
 
 | | NMR structure of OtTx1a - AMP in DPC micelles | | Descriptor: | Spiderine-1a | | Authors: | Nadezhdin, K, Romanovskaya, D, Sachkova, M, Vassilevski, A, Grishin, E, Kovalchuk, S, Arseniev, A. | | Deposit date: | 2015-10-05 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Modular toxin from the lynx spider Oxyopes takobius: Structure of spiderine domains in solution and membrane-mimicking environment.
Protein Sci., 26, 2017
|
|
2N86
 
 | | NMR structure of OtTx1a - ICK | | Descriptor: | Spiderine-1a | | Authors: | Nadezhdin, K, Romanovskaya, D, Sachkova, M, Vassilevski, A, Grishin, E, Kovalchuk, S, Arseniev, A. | | Deposit date: | 2015-10-05 | | Release date: | 2016-10-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Modular toxin from the lynx spider Oxyopes takobius: Structure of spiderine domains in solution and membrane-mimicking environment.
Protein Sci., 26, 2017
|
|
2MZG
 
 | | Purotoxin-2 NMR structure in DPC micelles | | Descriptor: | Purotoxin-2 | | Authors: | Nadezhdin, K, Oparin, P, Vassilevski, A, Grishin, E, Arseniev, A. | | Deposit date: | 2015-02-12 | | Release date: | 2016-04-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of purotoxin-2 from wolf spider: modular design and membrane-assisted mode of action in arachnid toxins.
Biochem. J., 473, 2016
|
|
7LQY
 
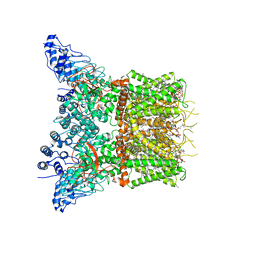 | | Structure of squirrel TRPV1 in apo state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoinositol, CHLORIDE ION, ... | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2021-02-15 | | Release date: | 2021-04-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Extracellular cap domain is an essential component of the TRPV1 gating mechanism.
Nat Commun, 12, 2021
|
|
8SI5
 
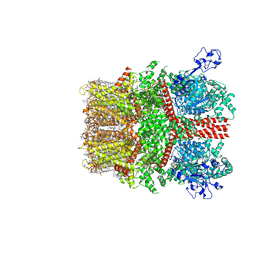 | | Cryo-EM structure of TRPM7 in MSP2N2 nanodisc in complex with agonist naltriben in open state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, (4bS,8R,8aS,14bR)-7-(cyclopropylmethyl)-5,6,7,8,9,14b-hexahydro-8aH-4,8-methanobis[1]benzofuro[3,2-e:2',3'-g]isoquinoline-1,8a-diol, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, ... | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-04-14 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.17 Å) | | Cite: | Structural mechanisms of TRPM7 activation and inhibition.
Nat Commun, 14, 2023
|
|
8SI6
 
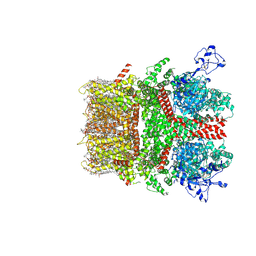 | | Cryo-EM structure of TRPM7 in MSP2N2 nanodisc in complex with agonist naltriben in closed state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, (4bS,8R,8aS,14bR)-7-(cyclopropylmethyl)-5,6,7,8,9,14b-hexahydro-8aH-4,8-methanobis[1]benzofuro[3,2-e:2',3'-g]isoquinoline-1,8a-diol, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, ... | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-04-14 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Structural mechanisms of TRPM7 activation and inhibition.
Nat Commun, 14, 2023
|
|
8SI4
 
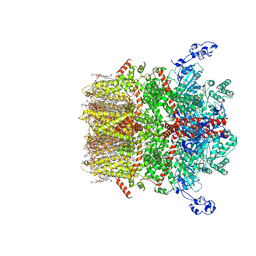 | | Cryo-EM structure of TRPM7 N1098Q mutant in GDN detergent in open state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, CALCIUM ION, ... | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-04-14 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Structural mechanisms of TRPM7 activation and inhibition.
Nat Commun, 14, 2023
|
|
8SI7
 
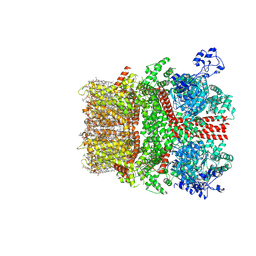 | | Cryo-EM structure of TRPM7 in GDN detergent in complex with inhibitor VER155008 in closed state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, 4-[[(2R,3S,4R,5R)-5-[6-amino-8-[(3,4-dichlorophenyl)methylamino]purin-9-yl]-3,4-dihydroxy-oxolan-2-yl]methoxymethyl]benzonitrile, ... | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-04-14 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structural mechanisms of TRPM7 activation and inhibition.
Nat Commun, 14, 2023
|
|
8SIA
 
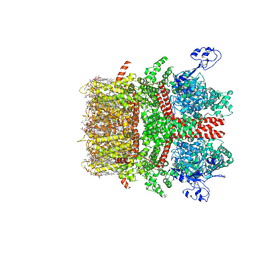 | | Cryo-EM structure of TRPM7 N1098Q mutant in GDN detergent in complex with inhibitor NS8593 in closed state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, CALCIUM ION, ... | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-04-14 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural mechanisms of TRPM7 activation and inhibition.
Nat Commun, 14, 2023
|
|
8SI2
 
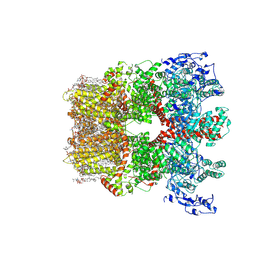 | | Cryo-EM structure of TRPM7 in MSP2N2 nanodisc in apo state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, CHOLESTEROL, ... | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-04-14 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Structural mechanisms of TRPM7 activation and inhibition.
Nat Commun, 14, 2023
|
|
8SI3
 
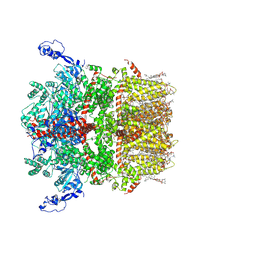 | | Cryo-EM structure of TRPM7 in GDN detergent in apo state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, CALCIUM ION, ... | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-04-14 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Structural mechanisms of TRPM7 activation and inhibition.
Nat Commun, 14, 2023
|
|
8SI8
 
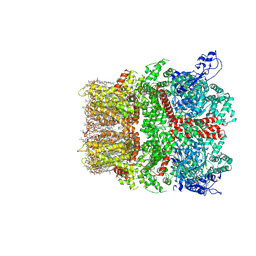 | | Cryo-EM structure of TRPM7 N1098Q mutant in GDN detergent in complex with inhibitor VER155008 in closed state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, 4-[[(2R,3S,4R,5R)-5-[6-amino-8-[(3,4-dichlorophenyl)methylamino]purin-9-yl]-3,4-dihydroxy-oxolan-2-yl]methoxymethyl]benzonitrile, ... | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-04-14 | | Release date: | 2023-05-17 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structural mechanisms of TRPM7 activation and inhibition.
Nat Commun, 14, 2023
|
|
8T1B
 
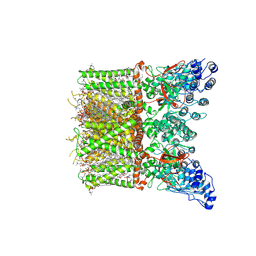 | | Cryo-EM structure of full-length human TRPV4 in apo state | | Descriptor: | (2R)-2-{[(4-O-hexopyranosyl-beta-D-glucopyranosyl)oxy]methyl}-4-{[(25R)-5beta,14beta,17beta-spirostan-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, SODIUM ION, Transient receptor potential cation channel subfamily V member 4/Enhanced green fluorescent protein chimera, ... | | Authors: | Nadezhdin, K.D, Talyzina, I.A, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-06-02 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of human TRPV4 in complex with GTPase RhoA.
Nat Commun, 14, 2023
|
|
2LLM
 
 | |
8SIB
 
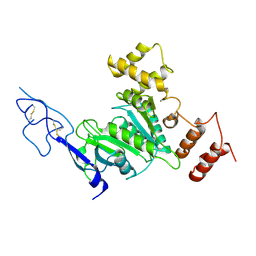 | |
8T1C
 
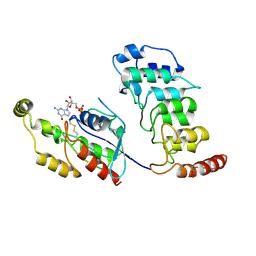 | | Cryo-EM structure of human TRPV4 ankyrin repeat domain in complex with GTPase RhoA | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Transforming protein RhoA, Transient receptor potential cation channel subfamily V member 4-Enhanced green fluorescent protein chimera | | Authors: | Nadezhdin, K.D, Talyzina, I.A, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-06-02 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structure of human TRPV4 in complex with GTPase RhoA.
Nat Commun, 14, 2023
|
|
7RAU
 
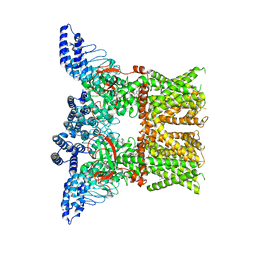 | | Structure of TRPV3 in complex with osthole | | Descriptor: | 7-methoxy-8-(3-methylbut-2-enyl)chromen-2-one, Transient receptor potential cation channel subfamily V member 3 | | Authors: | Neuberger, A, Nadezhdin, K.D, Sobolevsky, A.I. | | Deposit date: | 2021-07-02 | | Release date: | 2021-09-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Structural mechanism of TRPV3 channel inhibition by the plant-derived coumarin osthole.
Embo Rep., 22, 2021
|
|
7RAS
 
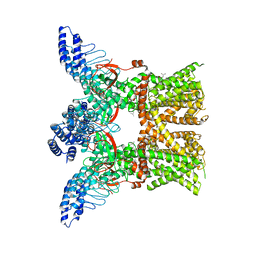 | | Structure of TRPV3 in complex with osthole | | Descriptor: | 7-methoxy-8-(3-methylbut-2-enyl)chromen-2-one, Transient receptor potential cation channel subfamily V member 3 | | Authors: | Neuberger, A, Nadezhdin, K.D, Sobolevsky, A.I. | | Deposit date: | 2021-07-02 | | Release date: | 2021-09-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structural mechanism of TRPV3 channel inhibition by the plant-derived coumarin osthole.
Embo Rep., 22, 2021
|
|
7UGG
 
 | | Cryo-EM structure of TRPV3 in complex with the anesthetic dyclonine | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Dyclonine, SODIUM ION, ... | | Authors: | Neuberger, A, Nadezhdin, K.D, Sobolevsky, A.I. | | Deposit date: | 2022-03-24 | | Release date: | 2022-06-08 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural mechanism of TRPV3 channel inhibition by the anesthetic dyclonine.
Nat Commun, 13, 2022
|
|
7D2K
 
 | | Crystal structure of rat TRPV6 in complex with (4- phenylcyclohexyl)piperazine inhibitor Br-cis-22a | | Descriptor: | 1-(5-bromanylpyridin-3-yl)-4-[4-(3-methylphenyl)cyclohexyl]piperazin-4-ium, CALCIUM ION, Transient receptor potential cation channel subfamily V member 6 | | Authors: | Singh, A.K, Neuberger, A, Nadezhdin, K.D, Sobolevsky, A.I. | | Deposit date: | 2020-09-16 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.698 Å) | | Cite: | Inactivation-mimicking block of the epithelial calcium channel TRPV6.
Sci Adv, 6, 2020
|
|
1ZNU
 
 | | Structure of cyclotide Kalata B1 in DPC micelles solution | | Descriptor: | Kalata B1 | | Authors: | Shenkarev, Z.O, Nadezhdin, K.D, Sobol, V.A, Sobol, A.G, Skjeldal, L, Arseniev, A.S. | | Deposit date: | 2005-05-12 | | Release date: | 2006-04-25 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | Conformation and mode of membrane interaction in cyclotides. Spatial structure of kalata B1 bound to a dodecylphosphocholine micelle.
Febs J., 273, 2006
|
|
