5AVH
 
 | | The 0.90 angstrom structure (I222) of glucose isomerase crystallized in high-strength agarose hydrogel | | Descriptor: | Xylose isomerase | | Authors: | Sugiyama, S, Shimizu, N, Maruyama, N, Sazaki, G, Adachi, H, Takano, K, Murakami, S, Inoue, T, Mori, Y, Matsumura, H. | | Deposit date: | 2015-06-16 | | Release date: | 2015-07-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Growth of protein crystals in hydrogels prevents osmotic shock
J.Am.Chem.Soc., 134, 2012
|
|
3U7F
 
 | | Crystal structure of mPNKP catalytic fragment (D170A) bound to single-stranded DNA (TCCTCp) | | Descriptor: | Bifunctional polynucleotide phosphatase/kinase, DNA, GLYCEROL, ... | | Authors: | Coquelle, N, Havali, Z, Bernstein, N, Green, R, Glover, J.N.M. | | Deposit date: | 2011-10-13 | | Release date: | 2011-12-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the phosphatase activity of polynucleotide kinase/phosphatase on single- and double-stranded DNA substrates.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6TNS
 
 | | PI3K delta in complex with 2methoxyN[2methoxy5(7{[(2R)4(oxetan3 yl)morpholin2yl]methoxy}1,3dihydro2 benzofuran5yl)pyridin3yl]ethane1 sulfonamide | | Descriptor: | 2-methoxy-~{N}-[2-methoxy-5-[7-[[(2~{R})-4-(oxetan-3-yl)morpholin-2-yl]methoxy]-1,3-dihydro-2-benzofuran-5-yl]pyridin-3-yl]ethanesulfonamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Convery, M.A, Rowland, P, Henley, Z.A, Barton, N, Down, K. | | Deposit date: | 2019-12-10 | | Release date: | 2020-01-01 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Optimization of Orally Bioavailable PI3K delta Inhibitors and Identification of Vps34 as a Key Selectivity Target.
J.Med.Chem., 63, 2020
|
|
4LE1
 
 | | Crystal structure of the receiver domain of DesR in the inactive state | | Descriptor: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Trajtenberg, F, Larrieux, N, Buschiazzo, A. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Allosteric activation of bacterial response regulators: the role of the cognate histidine kinase beyond phosphorylation.
MBio, 5, 2014
|
|
5AVN
 
 | | The 1.03 angstrom structure (P212121) of glucose isomerase crystallized in high-strength agarose hydrogel | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Sugiyama, S, Shimizu, N, Maruyama, N, Sazaki, G, Adachi, H, Takano, K, Murakami, S, Inoue, T, Mori, Y, Matsumura, H. | | Deposit date: | 2015-06-23 | | Release date: | 2015-07-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Growth of protein crystals in hydrogels prevents osmotic shock
J.Am.Chem.Soc., 134, 2012
|
|
5B40
 
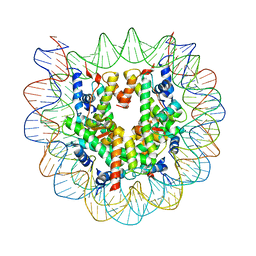 | | The nucleosome structure containing H2B-K120 and H4-K31 monoubiquitinations | | Descriptor: | DNA (146-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Machida, S, Sekine, S, Nishiyama, Y, Horikoshi, N, Kurumizaka, H. | | Deposit date: | 2016-03-22 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Monoubiquitination of histones H2B and H4 changes the nucleosome stability without affecting the nucleosome structure
To Be Published
|
|
5AXD
 
 | |
6T55
 
 | | Thrombin in Complex with Methylbenzylamine | | Descriptor: | 1-(4-methylphenyl)methanamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Ngo, K, Abazi, N, Heine, A, Klebe, G. | | Deposit date: | 2019-10-15 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Thrombin in Complex with Methylbenzylamine
To Be Published
|
|
6TS5
 
 | | Coagulation factor XI protease domain in complex with active site inhibitor | | Descriptor: | 2-[2-[3-[(3~{S})-3-azanyl-2,3-dihydro-1-benzofuran-5-yl]-5-propan-2-yl-phenyl]ethoxy]-3-methoxy-benzoic acid, Coagulation factor XI, DIMETHYL SULFOXIDE, ... | | Authors: | Renatus, M, Schiering, N. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Structure-Based Design and Preclinical Characterization of Selective and Orally Bioavailable Factor XIa Inhibitors: Demonstrating the Power of an Integrated S1 Protease Family Approach.
J.Med.Chem., 63, 2020
|
|
6TUB
 
 | | Beta-endorphin amyloid fibril | | Descriptor: | Beta-endorphin | | Authors: | Verasdonck, J, Seuring, C, Gath, J, Ghosh, D, Nespovitaya, N, Waelti, M.A, Maji, S, Cadalbert, R, Boeckmann, A, Guentert, P, Meier, B.H, Riek, R. | | Deposit date: | 2020-01-05 | | Release date: | 2020-10-28 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | The three-dimensional structure of human beta-endorphin amyloid fibrils.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5ARA
 
 | | Bovine mitochondrial ATP synthase state 1a | | Descriptor: | ATP SYNTHASE F(0) COMPLEX SUBUNIT B1, MITOCHONDRIAL, ATP SYNTHASE F(0) COMPLEX SUBUNIT C1, ... | | Authors: | Zhou, A, Rohou, A, Schep, D.G, Bason, J.V, Montgomery, M.G, Walker, J.E, Grigorieff, N, Rubinstein, J.L. | | Deposit date: | 2015-09-24 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM.
Elife, 4, 2015
|
|
6T8N
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K3007 | | Descriptor: | 1,2-ETHANEDIOL, Activin receptor type I, DIMETHYL SULFOXIDE, ... | | Authors: | Adamson, R.J, Williams, E.P, Bonomo, S, Rankin, S, Bacos, D, Rae, A, Cramp, S, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2019-10-24 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K3007
To Be Published
|
|
6T9P
 
 | | Human Butyrylcholinesterase in complex with 2-(N-hydroxyimino)-N-[(1R)-3-{4-[(2-methyl-1H-imidazol-1-yl)methyl]-1H-1,2,3-triazol-1-yl}-1- phenylpropyl]acetamide | | Descriptor: | (R,E)-2-(hydroxyimino)-N-(3-(4-((2-methyl-1H-imidazol-1-yl)methyl)-1H-1,2,3-triazol-1-yl)-1-phenylpropyl)acetamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[1-deoxy-alpha-D-tagatopyranose-(2-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, Sinko, G, Marakovic, N, Knezevic, A. | | Deposit date: | 2019-10-28 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Enantioseparation, in vitro testing, and structural characterization of triple-binding reactivators of organophosphate-inhibited cholinesterases.
Biochem.J., 477, 2020
|
|
5AZ3
 
 | | Crystal structure of heme binding protein HmuT | | Descriptor: | ABC-type transporter, periplasmic component, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Muraki, N, Aono, S. | | Deposit date: | 2015-09-18 | | Release date: | 2015-11-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.423 Å) | | Cite: | Structural Basis for Heme Recognition by HmuT Responsible for Heme Transport to the Heme Transporter in Corynebacterium glutamicum
Chem Lett., 2015
|
|
5AZD
 
 | | Crystal structure of thermophilic rhodopsin. | | Descriptor: | Bacteriorhodopsin | | Authors: | Mizutani, K, Hashimoto, N, Tsukamoto, T, Yamashita, K, Yamamoto, M, Sudo, Y, Murata, T. | | Deposit date: | 2015-09-30 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray crystallographic structure of thermophilic rhodopsin: implications for high thermal stability and optogenetic availability.
To Be Published
|
|
4M6Q
 
 | | Identification of Amides Derived From 1H-Pyrazolo[3,4-b]pyridine-5-carboxylic Acid as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | 1-(5-O-phosphono-beta-D-ribofuranosyl)-N-(4-{[3-(trifluoromethyl)phenyl]sulfonyl}benzyl)-1H-pyrazolo[3,4-b]pyridine-5-carboxamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION, ... | | Authors: | Giannetti, A.M, Zheng, X, Skelton, N, Wang, W, Bravo, B, Feng, Y, Gunzner-Toste, J, Ho, Y, Hua, R, Wang, C, Zhao, Q, Liederer, B.M, Liu, Y, O'Brien, T, Oeh, J, Sampath, D, Shen, Y, Wang, L, Wu, H, Xiao, Y, Yuen, P, Zak, M, Zhao, G, Dragovich, P.S. | | Deposit date: | 2013-08-10 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Identification of amides derived from 1H-pyrazolo[3,4-b]pyridine-5-carboxylic acid as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
5B1O
 
 | | DHp domain structure of EnvZ P248A mutant | | Descriptor: | Osmolarity sensor protein EnvZ | | Authors: | Okajima, T, Eguchi, Y, Tochio, N, Inukai, Y, Shimizu, R, Ueda, S, Shinya, S, Kigawa, T, Fukamizo, T, Igarashi, M, Utsumi, R. | | Deposit date: | 2015-12-09 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Angucycline antibiotic waldiomycin recognizes common structural motif conserved in bacterial histidine kinases
J. Antibiot., 70, 2017
|
|
4M6P
 
 | | Identification of Amides Derived From 1H-Pyrazolo[3,4-b]pyridine-5-carboxylic Acid as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | N-[4-(phenylsulfonyl)benzyl]-2H-pyrazolo[3,4-b]pyridine-5-carboxamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Giannetti, A.M, Zheng, X, Skelton, N, Wang, W, Bravo, B, Feng, Y, Gunzner-Toste, J, Ho, Y, Hua, R, Wang, C, Zhao, Q, Liederer, B.M, Liu, Y, O'Brien, T, Oeh, J, Sampath, D, Shen, Y, Wang, L, Wu, H, Xiao, Y, Yuen, P, Zak, M, Zhao, G, Dragovich, P.S. | | Deposit date: | 2013-08-10 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification of amides derived from 1H-pyrazolo[3,4-b]pyridine-5-carboxylic acid as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
5BRT
 
 | | Crystal Structure of 2-hydroxybiphenyl 3-monooxygenase from Pseudomonas azelaica with 2-hydroxybiphenyl in the active site | | Descriptor: | 2-HYDROXYBIPHENYL, 2-hydroxybiphenyl-3-monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kanteev, M, Bregman-Cohen, A, Deri, B, Adir, N, Fishman, A. | | Deposit date: | 2015-06-01 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A crystal structure of 2-hydroxybiphenyl 3-monooxygenase with bound substrate provides insights into the enzymatic mechanism.
Biochim.Biophys.Acta, 1854, 2015
|
|
4LUE
 
 | | Crystal Structure of HCK in complex with 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine (resulting from displacement of SKF86002) | | Descriptor: | 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Parker, L.J, Tanaka, A, Handa, N, Honda, K, Tomabechi, Y, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-25 | | Release date: | 2014-02-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Kinase crystal identification and ATP-competitive inhibitor screening using the fluorescent ligand SKF86002.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3NOE
 
 | | Crystal Structure of Dihydrodipicolinate synthase from Pseudomonas aeruginosa | | Descriptor: | Dihydrodipicolinate synthase, S-1,2-PROPANEDIOL | | Authors: | Kaur, N, Kumar, S, Singh, N, Gautam, A, Sharma, R, Sharma, S, Tewari, R, Singh, T.P. | | Deposit date: | 2010-06-25 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal Structure of Dihydrodipicolinate synthase from Pseudomonas aeruginosa
To be Published
|
|
4Z2L
 
 | | Serratia marcescens Chitinase B complexed with macrolide inhibitor 33 | | Descriptor: | (3R,4S,5S,6R,7R,9R,10S,11S,12R,13S,14R)-14-ethyl-7,10,12,13-tetrahydroxy-3,5,7,9,11,13-hexamethyl-2-oxo-6-(prop-2-yn-1-yloxy)oxacyclotetradecan-4-yl {3-[N'-(methylcarbamoyl)carbamimidamido]propyl}carbamate, CHLORIDE ION, Chitinase B, ... | | Authors: | Maita, N, Sugawara, A, Sunazuka, T. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Creation of Customized Bioactivity within a 14-Membered Macrolide Scaffold: Design, Synthesis, and Biological Evaluation Using a Family-18 Chitinase
J.Med.Chem., 58, 2015
|
|
4Z2G
 
 | | Serratia marcescens Chitinase B complexed with macrolide inhibitor 26 | | Descriptor: | (1R,2R,3R,6R,7S,8S,9R,10R,12R,13S,17S)-3-ethyl-2,10-dihydroxy-2,6,8,10,12,15,15,17-octamethyl-5-oxo-9-(prop-2-yn-1-yloxy)-4,14,16-trioxabicyclo[11.3.1]heptadec-7-yl {4-[N'-(methylcarbamoyl)carbamimidamido]butyl}carbamate, Chitinase B | | Authors: | Maita, N, Sugawara, A, Sunazuka, T. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Creation of Customized Bioactivity within a 14-Membered Macrolide Scaffold: Design, Synthesis, and Biological Evaluation Using a Family-18 Chitinase
J.Med.Chem., 58, 2015
|
|
6SWI
 
 | |
6SWL
 
 | | The REC domain of XynC, a response regulator from Geobacillus stearothermophilus | | Descriptor: | MAGNESIUM ION, Two-component response regulator | | Authors: | Lansky, S, Lavid, N, Shoham, Y, Shoham, G. | | Deposit date: | 2019-09-22 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | The REC domain of XynC, a response regulator from Geobacillus stearothermophilus
To Be Published
|
|
