5LLU
 
 | | Structure of the thermostabilized EAAT1 cryst-II mutant in complex with L-ASP | | Descriptor: | ASPARTIC ACID, Excitatory amino acid transporter 1,Neutral amino acid transporter B(0),Excitatory amino acid transporter 1, SODIUM ION | | Authors: | Canul-Tec, J, Assal, R, Legrand, P, Reyes, N. | | Deposit date: | 2016-07-28 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Structure and allosteric inhibition of excitatory amino acid transporter 1.
Nature, 544, 2017
|
|
8U47
 
 | |
5LM7
 
 | | Crystal structure of the lambda N-Nus factor complex | | Descriptor: | 30S ribosomal protein S10, Antitermination protein N, N utilization substance protein B homolog, ... | | Authors: | Said, N, Santos, K, Weber, G, Wahl, M.C. | | Deposit date: | 2016-07-29 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structural basis for lambda N-dependent processive transcription antitermination.
Nat Microbiol, 2, 2017
|
|
8GL3
 
 | |
8OSO
 
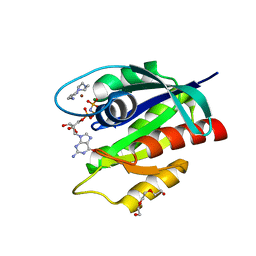 | | GTPase HRAS in complex with Zn-cyclen under 500 MPa pressure | | Descriptor: | 1,4,7,10-tetraazacyclododecane, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Colloc'h, N, Prange, T, Girard, E, Kalbitzer, H.R. | | Deposit date: | 2023-04-19 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High Pressure Promotes Binding of the Allosteric Inhibitor Zn 2+ -Cyclen in Crystals of Activated H-Ras.
Chemistry, 30, 2024
|
|
5LMZ
 
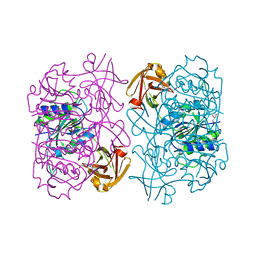 | | Fluorinase from Streptomyces sp. MA37 | | Descriptor: | 1-DEAZA-ADENOSINE, CHLORIDE ION, Fluorinase, ... | | Authors: | Mann, G, O'Hagan, D, Naismith, J.H, Bandaranayaka, N. | | Deposit date: | 2016-08-02 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Identification of fluorinases from Streptomyces sp MA37, Norcardia brasiliensis, and Actinoplanes sp N902-109 by genome mining.
Chembiochem, 15, 2014
|
|
8GKN
 
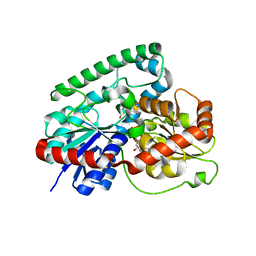 | | Crystal structure of TuUGT202A2 (Tetur22g00270) in complex with S-naringenin | | Descriptor: | NARINGENIN, UDP-glycosyltransferase 202A2, URIDINE-5'-DIPHOSPHATE | | Authors: | Arriaza, R.H, Dermauw, W, Wybouw, N, Van Leeuwen, T, Chruszcz, M. | | Deposit date: | 2023-03-20 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of TuUGT202A2 (Tetur22g00270) in complex with S-naringenin
To Be Published
|
|
8GSK
 
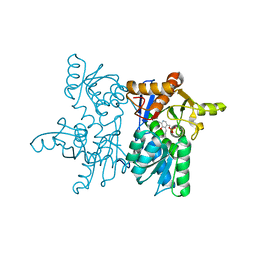 | |
7U8K
 
 | | Magic Angle Spinning NMR Structure of Human Cofilin-2 Assembled on Actin Filaments | | Descriptor: | Actin, alpha skeletal muscle, Cofilin-2 | | Authors: | Kraus, J, Russell, R, Kudryashova, E, Xu, C, Katyal, N, Kudryashov, D, Perilla, J.R, Polenova, T. | | Deposit date: | 2022-03-08 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-23 | | Method: | SOLID-STATE NMR | | Cite: | Magic angle spinning NMR structure of human cofilin-2 assembled on actin filaments reveals isoform-specific conformation and binding mode.
Nat Commun, 13, 2022
|
|
5ZK7
 
 | | Stapled-peptides tailored against initiation of translation | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, ACE-ARG-TYR-SER-ARG-MK8-GLN-LEU-LEU-MK8-LEU-PHE-ARG-NH2, CHLORIDE ION, ... | | Authors: | Lama, D, Liberator, A, Frosi, Y, Nakhle, J, Tsomia, N, Bashir, T, Lane, D.P, Brown, C.J, Verma, C.S, Auvin, S, Ciesielski, F, Uhring, M. | | Deposit date: | 2018-03-23 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural insights reveal a recognition feature for tailoring hydrocarbon stapled-peptides against the eukaryotic translation initiation factor 4E protein.
Chem Sci, 10, 2019
|
|
5ZML
 
 | | Stapled-peptides tailored against initiation of translation | | Descriptor: | 1,2-ETHANEDIOL, ACE-LYS-LYS-ARG-TYR-SER-ARG-MK8-GLN-LEU-LEU-MK8-PHE-ARG-ARG, Eukaryotic translation initiation factor 4E, ... | | Authors: | Lama, D, Liberator, A, Frosi, Y, Nakhle, J, Tsomia, N, Bashir, T, Lane, D.P, Brown, C.J, Verma, C.S, Auvin, S, Yano, J. | | Deposit date: | 2018-04-04 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights reveal a recognition feature for tailoring hydrocarbon stapled-peptides against the eukaryotic translation initiation factor 4E protein.
Chem Sci, 10, 2019
|
|
8Y4U
 
 | | Crystal structure of a His1 from oryza sativa | | Descriptor: | FE (III) ION, Fe(II)/2-oxoglutarate-dependent oxygenase | | Authors: | Wang, N, Ma, J.M, Shibing, H, Beibei, Y, He, Z, Dandan, L. | | Deposit date: | 2024-01-30 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of HPPD inhibitor sensitive protein from Oryza sativa.
Biochem.Biophys.Res.Commun., 704, 2024
|
|
8XMN
 
 | | Voltage-gated sodium channel Nav1.7 variant M2 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, N, Li, Z, Wu, Q, Huang, G. | | Deposit date: | 2023-12-27 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Dissection of the structure-function relationship of Na v channels.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5LKR
 
 | | Human Butyrylcholinesterase complexed with N-Propargyliperidines | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cholinesterase, ... | | Authors: | Coquelle, N, Knez, D, Colletier, J.P, Gobec, S. | | Deposit date: | 2016-07-23 | | Release date: | 2016-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | N-Propargylpiperidines with naphthalene-2-carboxamide or naphthalene-2-sulfonamide moieties: Potential multifunctional anti-Alzheimer's agents.
Bioorg. Med. Chem., 25, 2017
|
|
5LOB
 
 | |
5LOW
 
 | |
8XMO
 
 | | Voltage-gated sodium channel Nav1.7 variant M4 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, N, Li, Z, Wu, Q, Huang, G. | | Deposit date: | 2023-12-27 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Dissection of the structure-function relationship of Na v channels.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7VES
 
 | | Crystal Structure of nucleotide-free Irgb6 | | Descriptor: | T-cell-specific guanine nucleotide triphosphate-binding protein 2 | | Authors: | Saijo-Hamano, Y, Sakai, N, Nitta, R. | | Deposit date: | 2021-09-10 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of membrane recognition of Toxoplasma gondii vacuole by Irgb6.
Life Sci Alliance, 5, 2022
|
|
7VEX
 
 | | Crystal Structure of GTP-bound Irgb6 | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, T-cell-specific guanine nucleotide triphosphate-binding protein 2 | | Authors: | Saijo-Hamano, Y, Sakai, N, Nitta, R. | | Deposit date: | 2021-09-10 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural basis of membrane recognition of Toxoplasma gondii vacuole by Irgb6.
Life Sci Alliance, 5, 2022
|
|
5LZA
 
 | | Structure of the 70S ribosome with SECIS-mRNA and P-site tRNA (Initial complex, IC) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Bock, L.V, Maracci, C, Wang, Z, Paleskava, A, Konevega, A.L, Schroeder, G.F, Grubmueller, H, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The pathway to GTPase activation of elongation factor SelB on the ribosome.
Nature, 540, 2016
|
|
8XMM
 
 | | Voltage-gated sodium channel Nav1.7 variant M9 | | Descriptor: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Yan, N, Li, Z, Wu, Q, Huang, G. | | Deposit date: | 2023-12-27 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Dissection of the structure-function relationship of Na v channels.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5M4V
 
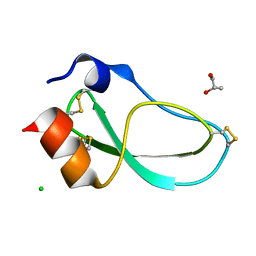 | | X-ray structure of the mambaquaretin-1, a selective antagonist of the vasopressin type 2 receptor | | Descriptor: | CHLORIDE ION, Mambaquaretin-1, S-1,2-PROPANEDIOL | | Authors: | Stura, E.A, Vera, L, Ciolek, J, Mourier, G, Gilles, N. | | Deposit date: | 2016-10-19 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Green mamba peptide targets type-2 vasopressin receptor against polycystic kidney disease.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5LCN
 
 | |
8V84
 
 | | 60S ribosome biogenesis intermediate (Dbp10 catalytic structure - Overall map) | | Descriptor: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 60S ribosomal protein L13-A, 60S ribosomal protein L14-A, ... | | Authors: | Cruz, V.E, Weirich, C.S, Peddada, N, Erzberger, J.P. | | Deposit date: | 2023-12-04 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The DEAD-box ATPase Dbp10/DDX54 initiates peptidyl transferase center formation during 60S ribosome biogenesis.
Nat Commun, 15, 2024
|
|
8W15
 
 | |
