6BA1
 
 | |
4IRB
 
 | | Crystal Structure of Vaccinia Virus Uracil DNA Glycosylase Mutant del171-172D4 | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Schormann, N, Zhukovskaya, N, Sartmatova, D, Nuth, M, Ricciardi, R.P, Chattopadhyay, D. | | Deposit date: | 2013-01-14 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mutations at the dimer interface affect both function and structure of the Vaccinia virus uracil DNA glycosylase
To be Published
|
|
1WFQ
 
 | | Solution structure of the first cold-shock domain of the human KIAA0885 protein (UNR protein) | | Descriptor: | UNR protein | | Authors: | Goroncy, A.K, Kigawa, T, Koshiba, S, Tomizawa, T, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-26 | | Release date: | 2004-11-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structures of the five constituent cold-shock domains (CSD) of the human UNR (upstream of N-ras) protein.
J.Struct.Funct.Genom., 11, 2010
|
|
6BBE
 
 | | Structure of N-glycosylated porcine surfactant protein-D | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | van Eijk, M, Rynkiewicz, M.J, Khatri, K, Leymarie, N, Zaia, J, White, M.R, Hartshorn, K.L, Cafarella, T.R, van Die, I, Hessing, M, Seaton, B.A, Haagsman, H.P. | | Deposit date: | 2017-10-18 | | Release date: | 2018-05-23 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Lectin-mediated binding and sialoglycans of porcine surfactant protein D synergistically neutralize influenza A virus.
J. Biol. Chem., 293, 2018
|
|
6BCK
 
 | | Crystal Structure of Broadly Neutralizing Antibody N49P7 in Complex with HIV-1 Clade AE strain 93TH057 gp120 core. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, N49P7 Fab heavy chain of N29P7 IgG, ... | | Authors: | Tolbert, W.D, Gohain, N, Pazgier, M. | | Deposit date: | 2017-10-20 | | Release date: | 2018-05-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of Near-Pan-neutralizing Antibodies against HIV-1 by Deconvolution of Plasma Humoral Responses.
Cell, 173, 2018
|
|
8UVZ
 
 | |
8UW0
 
 | |
4ITC
 
 | | Crystal Structure Analysis of the K1 Cleaved Adhesin domain of Lys-gingipain (Kgp) from Porphyromonas gingivalis W83 | | Descriptor: | CALCIUM ION, GLYCEROL, GUANIDINE, ... | | Authors: | Ganuelas, L.A, Li, N, Hunter, N, Collyer, C.A. | | Deposit date: | 2013-01-18 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The lysine gingipain adhesin domains from Porphyromonas gingivalis interact with erythrocytes and albumin: Structures correlate to function.
Eur J Microbiol Immunol (Bp), 3, 2013
|
|
6S8B
 
 | | Cryo-EM structure of the Type III-B Cmr-beta bound to cognate target RNA and AMPPnP, state 1 | | Descriptor: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | Authors: | Sofos, N, Montoya, G, Stella, S. | | Deposit date: | 2019-07-09 | | Release date: | 2020-07-08 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
6BKB
 
 | | Structure of Hepatitis C Virus Envelope Glycoprotein E2 core from genotype 6a bound to broadly neutralizing antibody AR3A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab AR3A heavy chain, ... | | Authors: | Tzarum, N, Wilson, I.A, Law, M. | | Deposit date: | 2017-11-08 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Genetic and structural insights into broad neutralization of hepatitis C virus by human VH1-69 antibodies.
Sci Adv, 5, 2019
|
|
1WWY
 
 | | Solution structure of the DUF1000 domain of a thioredoxin-like protein 1 | | Descriptor: | Thioredoxin-like protein 1 | | Authors: | Goroncy, A.K, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-18 | | Release date: | 2005-07-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal DUF1000 domain of the human thioredoxin-like 1 protein.
Proteins, 78, 2010
|
|
6BLG
 
 | | Crystal Structure of Sugar Transaminase from Klebsiella pneumoniae Complexed with PLP | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Maltseva, N, Kim, Y, Shatsman, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-10 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Crystal Structure of Sugar Transaminase from Klebsiella pneumoniae Complexed with PLP
To Be Published
|
|
6BH7
 
 | | Phosphotriesterase variant R18+254S | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase, ... | | Authors: | Miton, C.M, Campbell, E.C, Jackson, C.J, Tokuriki, N. | | Deposit date: | 2017-10-30 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Phosphotriesterase variant R18+254S
To Be Published
|
|
6BIC
 
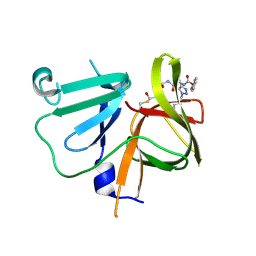 | | 2.25 A resolution structure of Norovirus 3CL protease in complex with a triazole-based macrocyclic inhibitor | | Descriptor: | (phenylmethyl) ~{N}-[(9~{S},12~{S},15~{S})-9-(hydroxymethyl)-12-(2-methylpropyl)-6,11,14-tris(oxidanylidene)-1,5,10,13,18,19-hexazabicyclo[15.2.1]icosa-17(20),18-dien-15-yl]carbamate, 3C-like protease | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Weerawarna, P.M, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | Deposit date: | 2017-11-01 | | Release date: | 2018-11-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Putative structural rearrangements associated with the interaction of macrocyclic inhibitors with norovirus 3CL protease.
Proteins, 87, 2019
|
|
6BKC
 
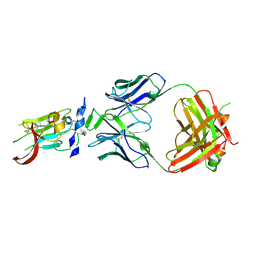 | | Structure of Hepatitis C Virus Envelope Glycoprotein E2 core from genotype 6a bound to broadly neutralizing antibody AR3B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab AR3B heavy chain, Fab AR3B light chain, ... | | Authors: | Tzarum, N, Wilson, I.A, Law, M. | | Deposit date: | 2017-11-08 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Genetic and structural insights into broad neutralization of hepatitis C virus by human VH1-69 antibodies.
Sci Adv, 5, 2019
|
|
6BHK
 
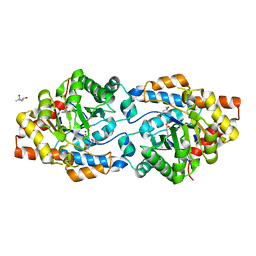 | | Phosphotriesterase variant R18deltaL7 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Phosphotriesterase, ZINC ION | | Authors: | Miton, C.M, Campbell, E.C, Jackson, C.J, Tokuriki, N. | | Deposit date: | 2017-10-30 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Phosphotriesterase variant R18deltaL7
To Be Published
|
|
8UQT
 
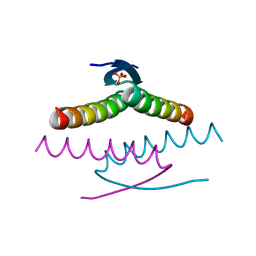 | | Crystal structure of the Tree Shrew p53 tetramerization domain | | Descriptor: | Cellular tumor antigen p53, SULFATE ION | | Authors: | Wahba, H.M, Sakaguchi, S, Nakagawa, N, Wada, J, Kamada, R, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2023-10-24 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Highly Similar Tetramerization Domains from the p53 Protein of Different Mammalian Species Possess Varying Biophysical, Functional and Structural Properties.
Int J Mol Sci, 24, 2023
|
|
6BIB
 
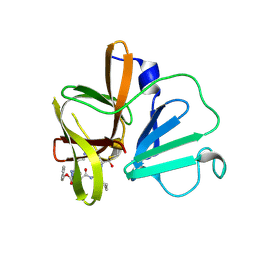 | | 1.95 A resolution structure of Norovirus 3CL protease in complex with a triazole-based macrocyclic inhibitor | | Descriptor: | 3C-like protease, benzyl [(9S,12S,15S)-12-(cyclohexylmethyl)-9-(hydroxymethyl)-6,11,14-trioxo-1,5,10,13,18,19-hexaazabicyclo[15.2.1]icosa-17(20),18-dien-15-yl]carbamate | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Weerawarna, P.M, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | Deposit date: | 2017-11-01 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Putative structural rearrangements associated with the interaction of macrocyclic inhibitors with norovirus 3CL protease.
Proteins, 87, 2019
|
|
2L5Z
 
 | | NMR structure of the A730 loop of the Neurospora VS ribozyme | | Descriptor: | RNA (26-MER) | | Authors: | Desjardins, G, Bonneau, E, Girard, N, Boisbouvier, J, Legault, P. | | Deposit date: | 2010-11-10 | | Release date: | 2011-02-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the A730 loop of the Neurospora VS ribozyme: insights into the formation of the active site.
Nucleic Acids Res., 39, 2011
|
|
8UQS
 
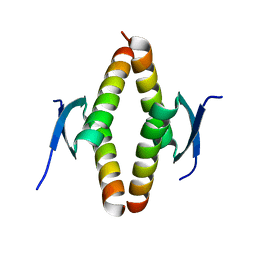 | | Crystal structure of the Opossum p53 tetramerization domain | | Descriptor: | Cellular tumor antigen p53 (Fragment) | | Authors: | Wahba, H.M, Sakaguchi, S, Nakagawa, N, Wada, J, Kamada, R, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2023-10-24 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Highly Similar Tetramerization Domains from the p53 Protein of Different Mammalian Species Possess Varying Biophysical, Functional and Structural Properties.
Int J Mol Sci, 24, 2023
|
|
8UQR
 
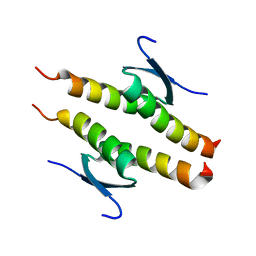 | | Crystal structure of the human p53 tetramerization domain | | Descriptor: | Cellular tumor antigen p53 | | Authors: | Wahba, H.M, Sakaguchi, S, Nakagawa, N, Wada, J, Kamada, R, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2023-10-24 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Highly Similar Tetramerization Domains from the p53 Protein of Different Mammalian Species Possess Varying Biophysical, Functional and Structural Properties.
Int J Mol Sci, 24, 2023
|
|
8UPT
 
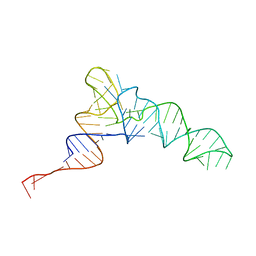 | | Candidatus Methanomethylophilus alvus tRNAPyl in A-site of ribosome | | Descriptor: | RNA (71-MER) | | Authors: | Krahn, N, Zhang, J, Melnikov, S.V, Tharp, J.M, Villa, A, Patel, A, Howard, R.J, Gabir, H, Patel, T.R, Stetefeld, J, Puglisi, J, Soll, D. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | tRNA shape is an identity element for an archaeal pyrrolysyl-tRNA synthetase from the human gut.
Nucleic Acids Res., 52, 2024
|
|
6BMF
 
 | | Vps4p-Vta1p complex with peptide binding to the central pore of Vps4p | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Han, H, Monroe, N, Shen, P, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2017-11-14 | | Release date: | 2017-12-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The AAA ATPase Vps4 binds ESCRT-III substrates through a repeating array of dipeptide-binding pockets.
Elife, 6, 2017
|
|
6BOZ
 
 | | Structure of human SETD8 in complex with covalent inhibitor MS4138 | | Descriptor: | 1,2-ETHANEDIOL, N-(3-{[7-(2-aminoethoxy)-6-methoxy-2-(pyrrolidin-1-yl)quinazolin-4-yl]amino}propyl)prop-2-enamide, N-lysine methyltransferase KMT5A | | Authors: | Babault, N, Anqi, M, Jin, J. | | Deposit date: | 2017-11-21 | | Release date: | 2019-05-01 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The dynamic conformational landscape of the protein methyltransferase SETD8.
Elife, 8, 2019
|
|
1WTN
 
 | | The structure of HEW Lysozyme Orthorhombic Crystal Growth under a High Magnetic Field | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Saijo, S, Yamada, Y, Sato, T, Tanaka, N, Matsui, T, Sazaki, G, Nakajima, K, Matsuura, Y. | | Deposit date: | 2004-11-25 | | Release date: | 2004-12-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Structural consequences of hen egg-white lysozyme orthorhombic crystal growth in a high magnetic field: validation of X-ray diffraction intensity, conformational energy searching and quantitative analysis of B factors and mosaicity.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
